"superoxide dismutase 1, soluble"
ZFIN 
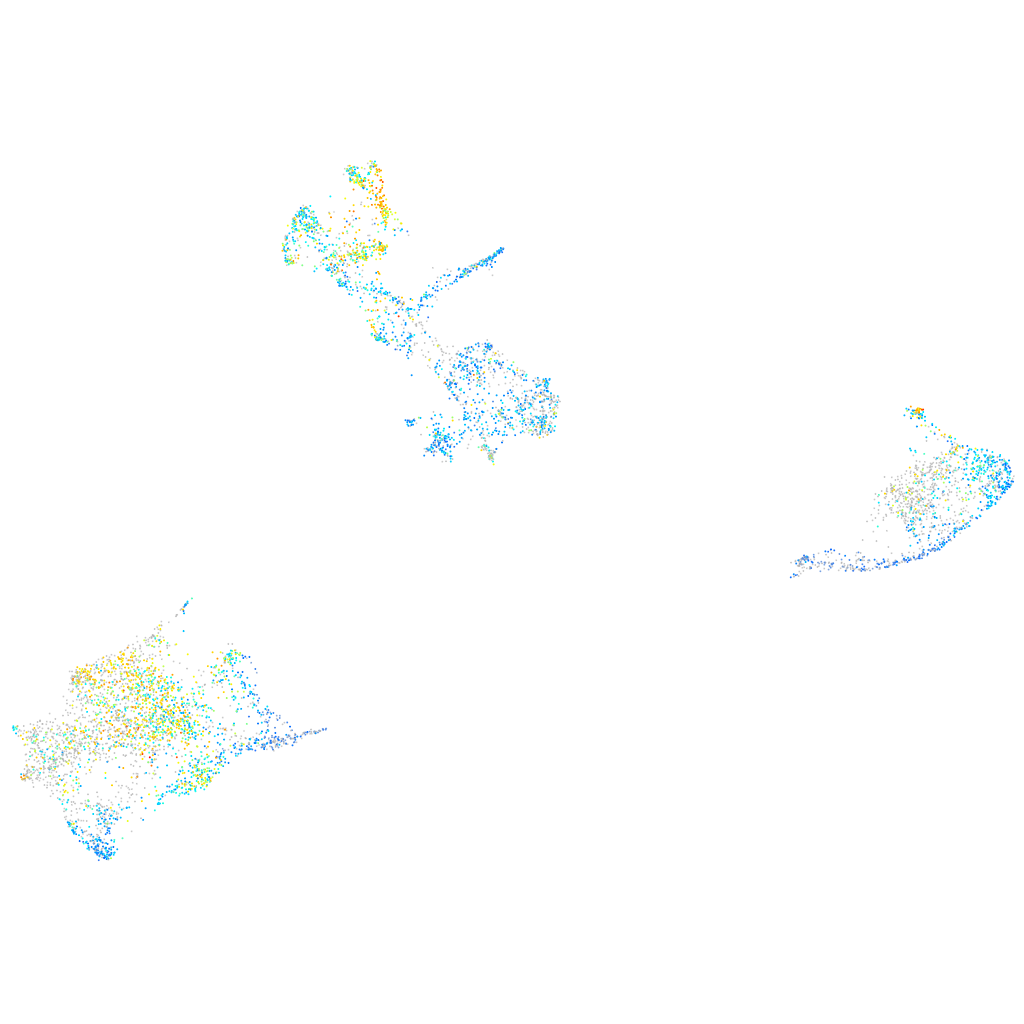
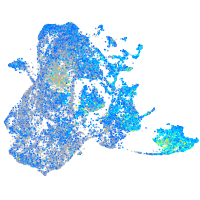
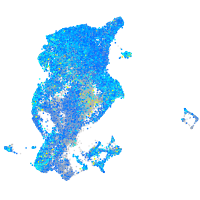
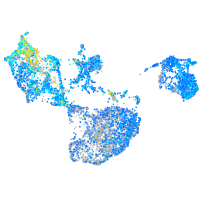
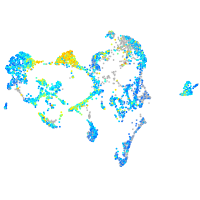
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| impdh1b | 0.279 | icn | -0.185 |
| prps1a | 0.274 | pmela | -0.163 |
| unm-sa821 | 0.274 | tyrp1b | -0.162 |
| si:dkey-197i20.6 | 0.274 | tyrp1a | -0.150 |
| glulb | 0.274 | dct | -0.147 |
| paics | 0.269 | zgc:91968 | -0.145 |
| zgc:110789 | 0.269 | s100a10b | -0.141 |
| lypc | 0.265 | tyr | -0.137 |
| gpnmb | 0.262 | oca2 | -0.134 |
| si:ch211-256m1.8 | 0.262 | anxa1a | -0.115 |
| apoda.1 | 0.257 | prkar1b | -0.109 |
| psat1 | 0.256 | slc24a5 | -0.107 |
| tagln3b | 0.255 | cdkn1ca | -0.107 |
| akap12a | 0.254 | slc1a3a | -0.106 |
| ponzr1 | 0.253 | msx1b | -0.102 |
| nme4 | 0.252 | tuba1a | -0.098 |
| pltp | 0.252 | tmem119b | -0.095 |
| atic | 0.251 | si:ch73-389b16.1 | -0.095 |
| defbl1 | 0.250 | ednrab | -0.094 |
| gmps | 0.241 | mitfa | -0.090 |
| sytl2b | 0.240 | si:ch211-222l21.1 | -0.086 |
| phyhd1 | 0.239 | mtbl | -0.086 |
| slc2a15a | 0.238 | lamp1a | -0.086 |
| tpd52l1 | 0.236 | tfap2a | -0.085 |
| guk1a | 0.231 | nr2f5 | -0.085 |
| slc25a36a | 0.231 | marcksb | -0.084 |
| mdh1aa | 0.231 | dio3a | -0.082 |
| si:ch73-89b15.3 | 0.229 | mical3b | -0.082 |
| phgdh | 0.229 | nova2 | -0.080 |
| fhl2b | 0.228 | tmeff1b | -0.079 |
| CT737162.3 | 0.227 | elavl3 | -0.079 |
| gapdhs | 0.227 | kita | -0.079 |
| clec3ba | 0.225 | tfap2e | -0.078 |
| fhl3a | 0.223 | gpm6aa | -0.078 |
| fhl2a | 0.223 | gstt1a | -0.077 |