si:ch73-304f21.1
ZFIN 
Other cell groups


all cells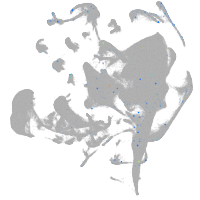 |
blastomeres |
PGCs |
endoderm |
axial mesoderm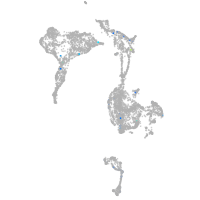 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia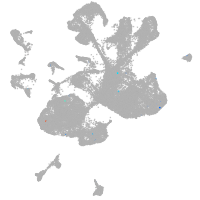 |
eye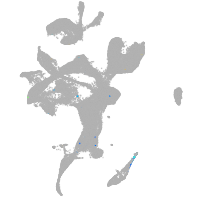 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| capn3a | 0.119 | gpm6aa | -0.020 |
| crygm2d4 | 0.113 | hmgb1b | -0.020 |
| crygm2d5 | 0.112 | nova2 | -0.020 |
| crygm2d18 | 0.110 | tuba1c | -0.020 |
| BX548032.3 | 0.106 | elavl3 | -0.019 |
| crygm2d21 | 0.106 | hmgb3a | -0.019 |
| crygm2d10 | 0.105 | stmn1b | -0.019 |
| crygm2d12 | 0.104 | ckbb | -0.018 |
| crygm2d3 | 0.103 | gpm6ab | -0.018 |
| crygmxl2 | 0.103 | si:dkey-276j7.1 | -0.018 |
| BX545855.1 | 0.101 | chd4a | -0.017 |
| crygm2d7 | 0.101 | marcksb | -0.017 |
| bfsp2 | 0.098 | mdkb | -0.017 |
| cryba1l2 | 0.098 | si:dkey-56m19.5 | -0.017 |
| crygm2d13 | 0.095 | tuba1a | -0.017 |
| crygm2d1 | 0.093 | zc4h2 | -0.017 |
| crygm2d20 | 0.093 | rnasekb | -0.016 |
| endou2 | 0.092 | si:ch211-137a8.4 | -0.016 |
| crygm2d8 | 0.090 | si:ch211-288g17.3 | -0.016 |
| mipa | 0.086 | tubb5 | -0.016 |
| crygm2d2 | 0.085 | gng3 | -0.015 |
| lim2.5 | 0.085 | hnrnpa0a | -0.015 |
| lim2.4 | 0.084 | nucks1a | -0.015 |
| crybb1l3 | 0.080 | rtn1b | -0.015 |
| lim2.3 | 0.080 | tmeff1b | -0.015 |
| cx23 | 0.078 | tmsb | -0.015 |
| gpx9 | 0.078 | cspg5a | -0.014 |
| crybb1l1 | 0.077 | cyt1 | -0.014 |
| crybb1l2 | 0.077 | fez1 | -0.014 |
| lim2.1 | 0.076 | krt4 | -0.014 |
| prx | 0.075 | myt1a | -0.014 |
| BFSP1 | 0.074 | myt1b | -0.014 |
| xirp1 | 0.074 | sncb | -0.014 |
| cryba2a | 0.073 | sox11b | -0.014 |
| mipb | 0.073 | atp6v0cb | -0.013 |




