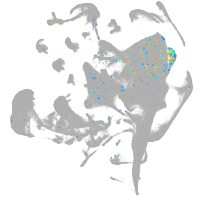
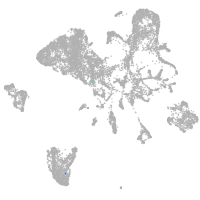
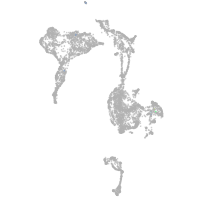
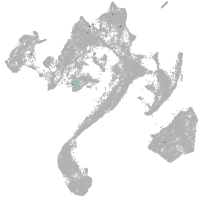
si:ch211-248l17.3
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glra4b | 0.241 | id1 | -0.034 |
| si:ch211-129p13.1 | 0.219 | sdc4 | -0.032 |
| pax6b | 0.172 | vamp3 | -0.032 |
| tkta | 0.170 | aldob | -0.031 |
| pax6a | 0.168 | bzw1b | -0.031 |
| zgc:92912 | 0.157 | tuba8l | -0.030 |
| calb2a | 0.157 | msna | -0.029 |
| pcbp3 | 0.148 | ccng1 | -0.028 |
| pax10 | 0.148 | eef1da | -0.028 |
| gria3a | 0.143 | eif4ebp3l | -0.028 |
| cabp1b | 0.141 | pcna | -0.028 |
| cplx3b | 0.140 | selenoh | -0.027 |
| mir181b-3 | 0.138 | si:ch211-286b5.5 | -0.027 |
| hmx4 | 0.137 | cd63 | -0.026 |
| lrtm1 | 0.137 | epcam | -0.026 |
| si:ch73-290k24.5 | 0.137 | COX7A2 (1 of many) | -0.026 |
| rbpms2a | 0.131 | ccnd1 | -0.025 |
| adgra1a | 0.129 | hdlbpa | -0.025 |
| rbpms2b | 0.125 | rps27l | -0.025 |
| pou4f3 | 0.124 | banf1 | -0.024 |
| XLOC-020666 | 0.123 | chaf1a | -0.024 |
| plppr4b | 0.123 | f11r.1 | -0.024 |
| BX005048.1 | 0.122 | lig1 | -0.024 |
| hmx1 | 0.120 | nutf2l | -0.024 |
| zgc:158291 | 0.120 | ptbp1a | -0.024 |
| isl2b | 0.119 | sox3 | -0.024 |
| neurod2 | 0.118 | tuba8l2 | -0.024 |
| LOC101886114 | 0.117 | ucp2 | -0.024 |
| ahr1b | 0.116 | cdca7b | -0.023 |
| rorb | 0.115 | eno3 | -0.023 |
| tfap2a | 0.114 | hspb1 | -0.023 |
| tfap2b | 0.114 | slc38a5b | -0.023 |
| anks1ab | 0.113 | her15.1 | -0.023 |
| snap25b | 0.110 | ahnak | -0.022 |
| cpne8 | 0.109 | akap12b | -0.022 |