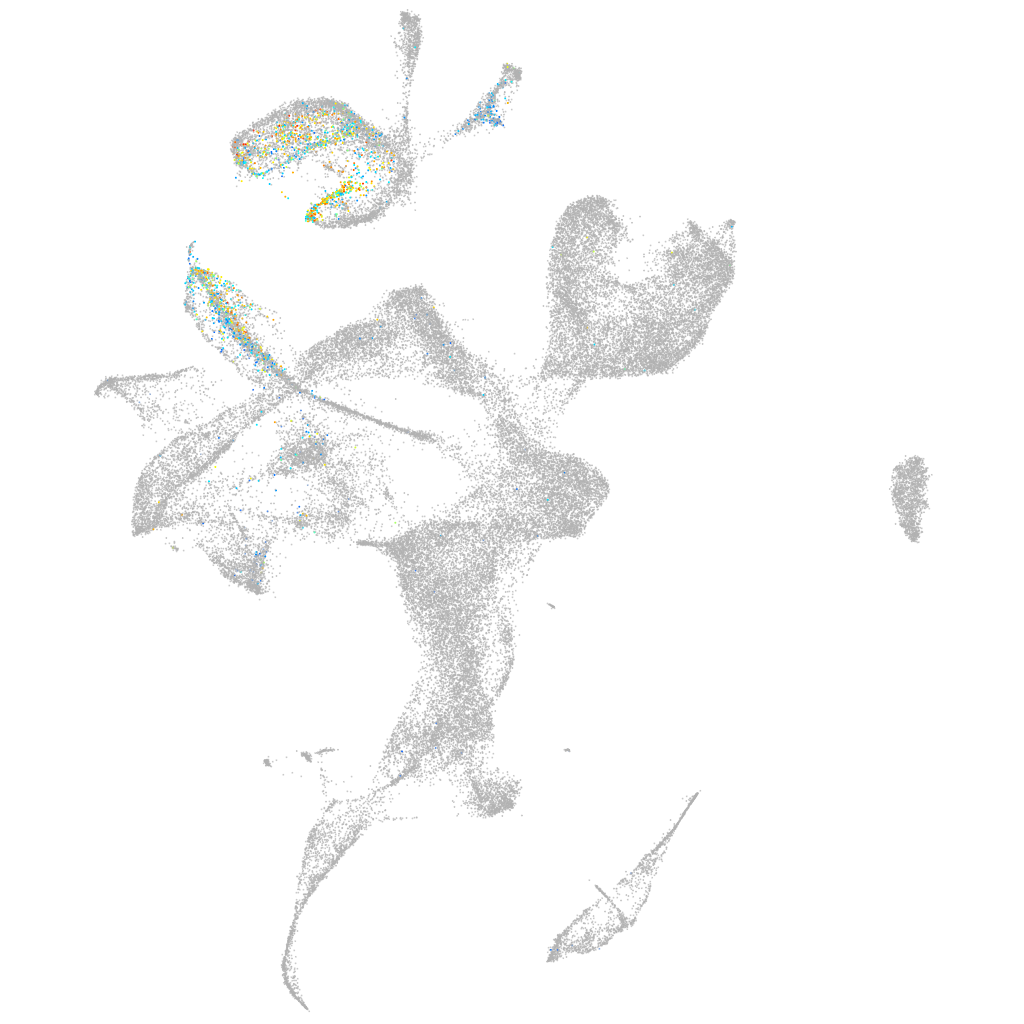si:ch211-248l17.3
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| calb2a | 0.358 | hmgb2a | -0.175 |
| neurod2 | 0.315 | msi1 | -0.100 |
| syt2a | 0.280 | pcna | -0.098 |
| zgc:92912 | 0.272 | mdka | -0.095 |
| tfap2a | 0.257 | msna | -0.095 |
| glra4b | 0.255 | fabp7a | -0.092 |
| mir181b-3 | 0.254 | ahcy | -0.090 |
| rbfox1 | 0.252 | neurod4 | -0.089 |
| elavl3 | 0.240 | stmn1a | -0.089 |
| eno2 | 0.239 | dut | -0.088 |
| rtn1b | 0.238 | tuba8l4 | -0.088 |
| si:ch73-290k24.5 | 0.237 | rrm1 | -0.088 |
| syt3 | 0.234 | crx | -0.087 |
| stxbp1a | 0.231 | eef1da | -0.087 |
| tfap2b | 0.229 | nutf2l | -0.087 |
| XLOC-020666 | 0.229 | tuba8l | -0.086 |
| ppfia4 | 0.223 | her15.1 | -0.085 |
| stx1b | 0.223 | ccnd1 | -0.085 |
| gria3a | 0.217 | otx5 | -0.085 |
| si:ch211-129p13.1 | 0.217 | selenoh | -0.084 |
| ywhag2 | 0.216 | mki67 | -0.083 |
| tkta | 0.212 | chaf1a | -0.082 |
| gng3 | 0.208 | syt5b | -0.081 |
| pax6a | 0.208 | id1 | -0.080 |
| ppp1r14ba | 0.200 | mcm7 | -0.080 |
| cplx2l | 0.200 | histh1l | -0.080 |
| atp2b3b | 0.197 | dek | -0.079 |
| adgra1a | 0.197 | ccna2 | -0.079 |
| cabp1b | 0.195 | lbr | -0.078 |
| sncb | 0.195 | ddah2 | -0.078 |
| zgc:153426 | 0.194 | COX7A2 (1 of many) | -0.078 |
| anks1ab | 0.194 | rrm2 | -0.076 |
| pax6b | 0.193 | cks1b | -0.076 |
| gabrb4 | 0.190 | lig1 | -0.075 |
| LOC101886114 | 0.190 | fen1 | -0.075 |