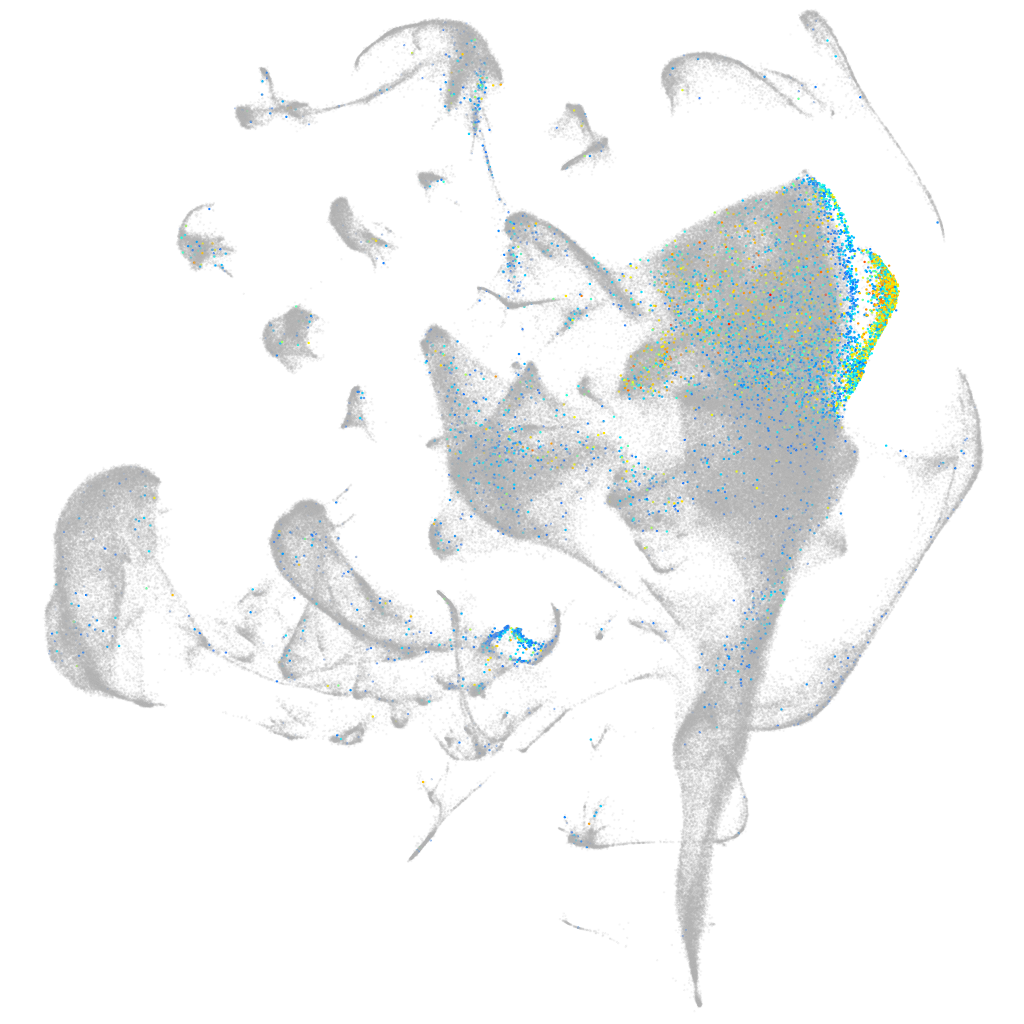zgc:158291
ZFIN 
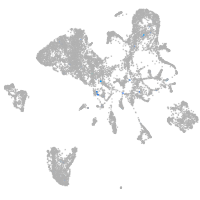
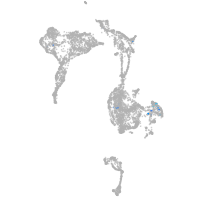
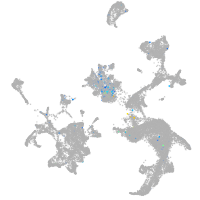
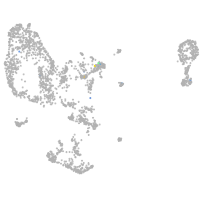
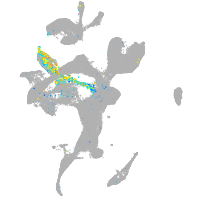
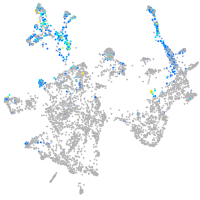
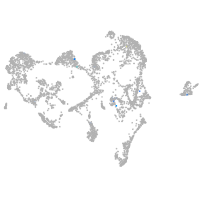
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rbpms2b | 0.398 | id1 | -0.075 |
| isl2b | 0.355 | sdc4 | -0.069 |
| rbpms2a | 0.292 | tuba8l | -0.067 |
| pou4f1 | 0.276 | qkia | -0.065 |
| pou4f3 | 0.255 | vamp3 | -0.065 |
| gpr132b | 0.246 | aldob | -0.062 |
| pou4f2 | 0.244 | selenoh | -0.058 |
| si:ch211-12e13.12 | 0.227 | ccnd1 | -0.056 |
| si:ch211-129p13.1 | 0.224 | ccng1 | -0.056 |
| barhl1a | 0.221 | pcna | -0.056 |
| cacnb3b | 0.201 | eif4ebp3l | -0.055 |
| CU862020.1 | 0.200 | hdlbpa | -0.055 |
| islr2 | 0.192 | msna | -0.055 |
| kif26bb | 0.189 | bzw1b | -0.054 |
| pou6f2 | 0.180 | cd63 | -0.054 |
| ebf3a | 0.180 | eef1da | -0.054 |
| pcbp4 | 0.174 | fosab | -0.053 |
| hmx4 | 0.171 | banf1 | -0.051 |
| inab | 0.165 | chaf1a | -0.051 |
| irx4a | 0.165 | nutf2l | -0.051 |
| si:dkey-280e21.3 | 0.165 | COX7A2 (1 of many) | -0.051 |
| elavl3 | 0.164 | atp1b1a | -0.050 |
| gpm6ab | 0.164 | zgc:162730 | -0.050 |
| hmx1 | 0.163 | junba | -0.049 |
| tfap2d | 0.163 | npm1a | -0.049 |
| scrt2 | 0.162 | zfp36l1a | -0.049 |
| pax6a | 0.156 | ahnak | -0.048 |
| mllt11 | 0.153 | slc38a5b | -0.048 |
| stmn1b | 0.151 | sparc | -0.048 |
| stmn2b | 0.151 | tuba8l2 | -0.048 |
| isl1 | 0.149 | dkc1 | -0.046 |
| mab21l1 | 0.149 | GCA | -0.046 |
| nrn1a | 0.148 | ier2a | -0.046 |
| barhl2 | 0.145 | mki67 | -0.046 |
| BX247868.1 | 0.141 | nop58 | -0.046 |