ring finger protein 32
ZFIN 
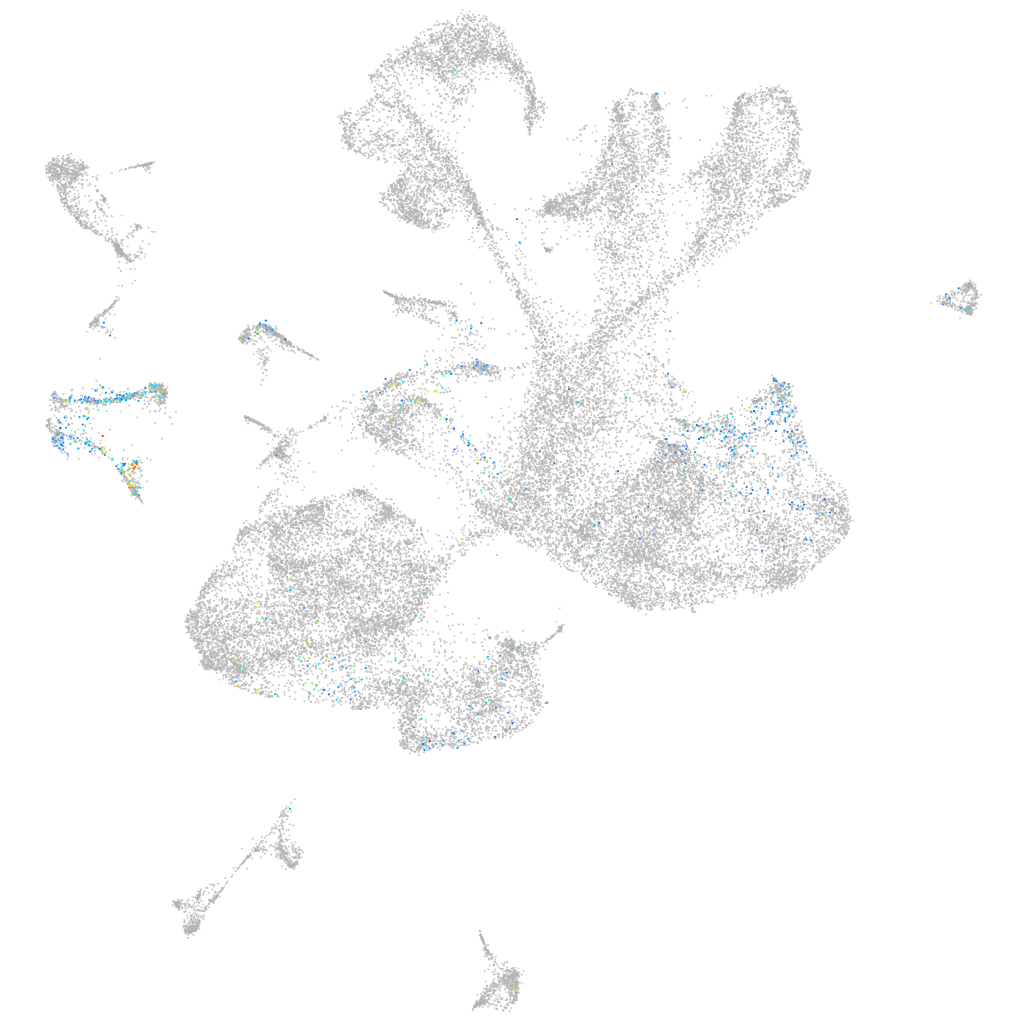
Other cell groups


all cells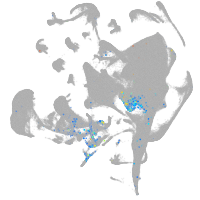 |
blastomeres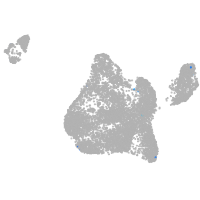 |
PGCs |
endoderm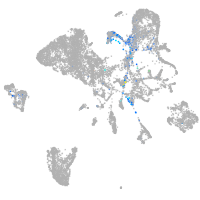 |
axial mesoderm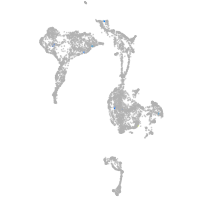 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia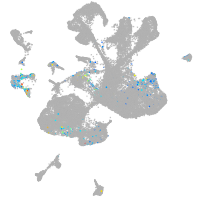 |
eye |
taste / olfactory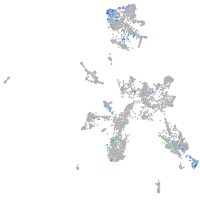 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slit1b | 0.259 | elavl3 | -0.072 |
| clu | 0.258 | ptmab | -0.070 |
| shhb | 0.244 | tmsb4x | -0.062 |
| shha | 0.238 | stmn1b | -0.062 |
| spon1b | 0.235 | ptmaa | -0.059 |
| arxa | 0.227 | hnrnpaba | -0.059 |
| myl9a | 0.226 | myt1b | -0.058 |
| vasna | 0.224 | tmsb | -0.058 |
| npr3 | 0.210 | hmgb3a | -0.057 |
| ntn1b | 0.202 | rtn1a | -0.055 |
| trabd2b | 0.202 | si:dkey-276j7.1 | -0.053 |
| ctgfa | 0.201 | tubb2b | -0.049 |
| cyr61l2 | 0.195 | si:ch73-386h18.1 | -0.049 |
| ptgs2a | 0.190 | tmeff1b | -0.048 |
| slit2 | 0.190 | FO082781.1 | -0.048 |
| enkur | 0.189 | gpm6ab | -0.048 |
| si:ch211-155m12.5 | 0.186 | si:dkey-56m19.5 | -0.048 |
| hpgd | 0.183 | stxbp1a | -0.046 |
| si:dkey-208c12.2 | 0.182 | nova1 | -0.046 |
| tbata | 0.181 | hmgb1b | -0.046 |
| foxa1 | 0.177 | pax3a | -0.046 |
| cavin2b | 0.176 | sox11b | -0.046 |
| loxl5b | 0.176 | ywhah | -0.045 |
| daw1 | 0.168 | gng3 | -0.045 |
| zgc:162730 | 0.168 | elavl4 | -0.045 |
| foxj1a | 0.166 | myt1a | -0.045 |
| si:ch211-163l21.7 | 0.165 | stx1b | -0.044 |
| tmem88b | 0.164 | marcksb | -0.044 |
| scospondin | 0.164 | scrt2 | -0.044 |
| col8a1a | 0.164 | gng2 | -0.043 |
| lcn15 | 0.163 | sncb | -0.043 |
| bicc2 | 0.163 | ckbb | -0.043 |
| ccdc114 | 0.163 | gas1b | -0.043 |
| pacrg | 0.162 | rtn4b | -0.043 |
| emp2 | 0.162 | cirbpb | -0.042 |