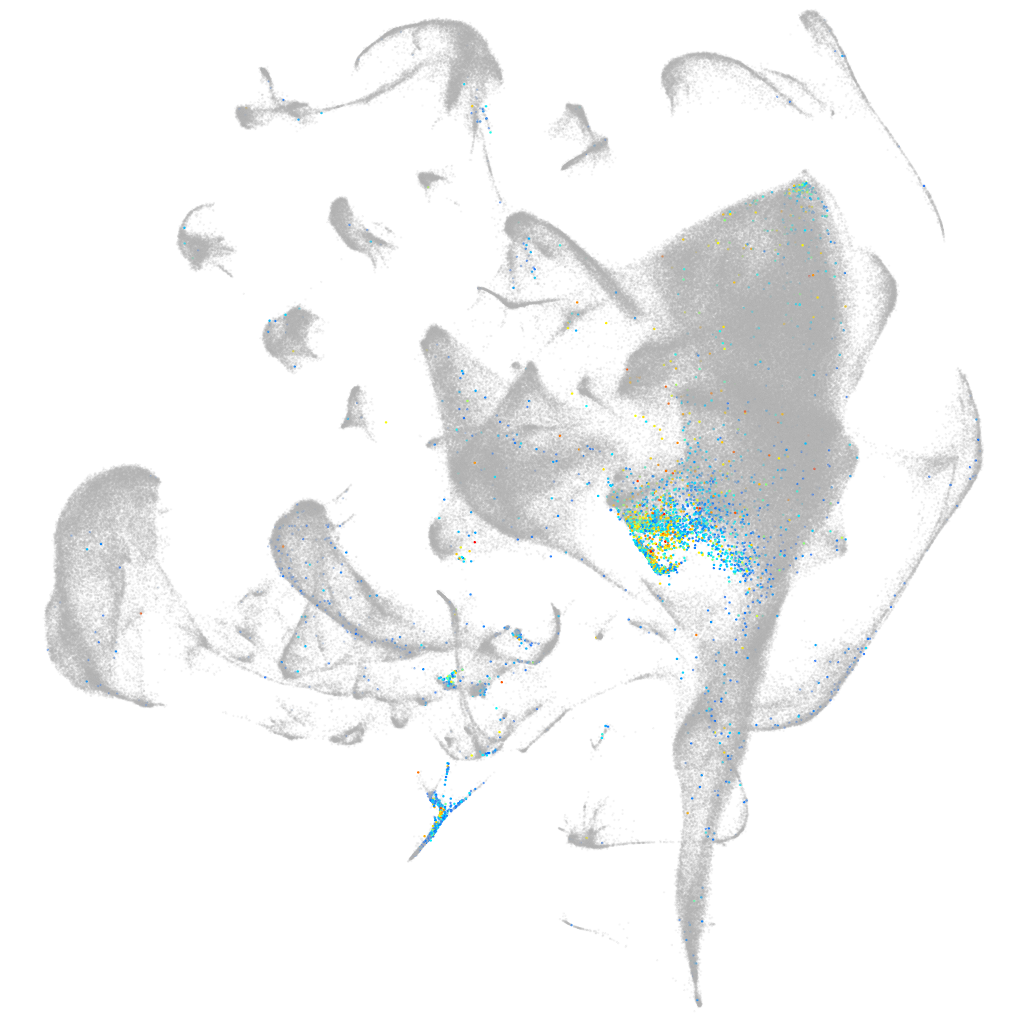
si:ch211-155m12.5
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm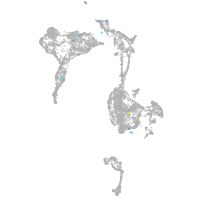 |
muscle 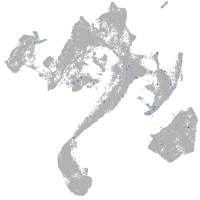 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 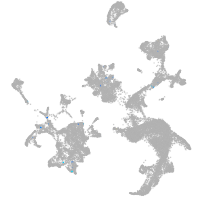 |
pronephros  |
neural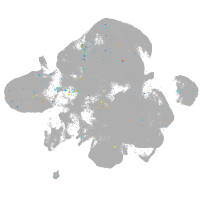 |
spinal cord / glia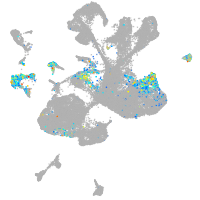 |
eye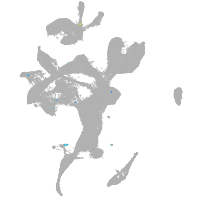 |
taste / olfactory |
otic / lateral line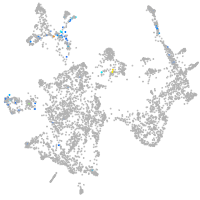 |
epidermis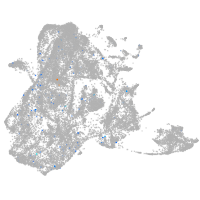 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.422 | stmn1b | -0.027 |
| tbata | 0.420 | celf2 | -0.024 |
| foxj1a | 0.379 | mab21l1 | -0.024 |
| enkur | 0.366 | scrt2 | -0.024 |
| smkr1 | 0.361 | wu:fb18f06 | -0.023 |
| cfap126 | 0.359 | cfl1l | -0.022 |
| morn3 | 0.356 | cyt1 | -0.022 |
| pacrg | 0.349 | fxyd6l | -0.022 |
| si:ch211-163l21.7 | 0.345 | hrc | -0.022 |
| si:dkey-148f10.4 | 0.341 | lye | -0.022 |
| ccdc169 | 0.336 | mgst1.2 | -0.022 |
| si:dkey-27p23.3 | 0.336 | pnp5a | -0.022 |
| ak7a | 0.332 | rbfox1 | -0.022 |
| spag6 | 0.330 | si:ch211-105c13.3 | -0.022 |
| ttc25 | 0.328 | zgc:193505 | -0.022 |
| cfap157 | 0.326 | arhgdig | -0.021 |
| ccdc151 | 0.324 | elavl4 | -0.021 |
| ccdc173 | 0.322 | FO082781.1 | -0.021 |
| LOC100329490 | 0.319 | icn2 | -0.021 |
| si:dkeyp-110a12.4 | 0.319 | krtt1c19e | -0.021 |
| dnali1 | 0.315 | mab21l2 | -0.021 |
| spata18 | 0.315 | scel | -0.021 |
| ccdc114 | 0.311 | snap25b | -0.021 |
| nme5 | 0.301 | ebf3a | -0.021 |
| rab36 | 0.296 | agr1 | -0.020 |
| dydc2 | 0.293 | caspb | -0.020 |
| nme8 | 0.288 | col1a2 | -0.020 |
| bicc2 | 0.287 | krt5 | -0.020 |
| spaca9 | 0.286 | ndrg4 | -0.020 |
| cfap52 | 0.283 | nrn1a | -0.020 |
| pih1d3 | 0.282 | ppl | -0.020 |
| si:ch211-163l21.10 | 0.282 | si:ch211-125o16.4 | -0.020 |
| lrrc51 | 0.280 | si:ch211-157c3.4 | -0.020 |
| lrrc74b | 0.277 | si:ch211-195b11.3 | -0.020 |
| ribc2 | 0.277 | si:ch73-52f15.5 | -0.020 |