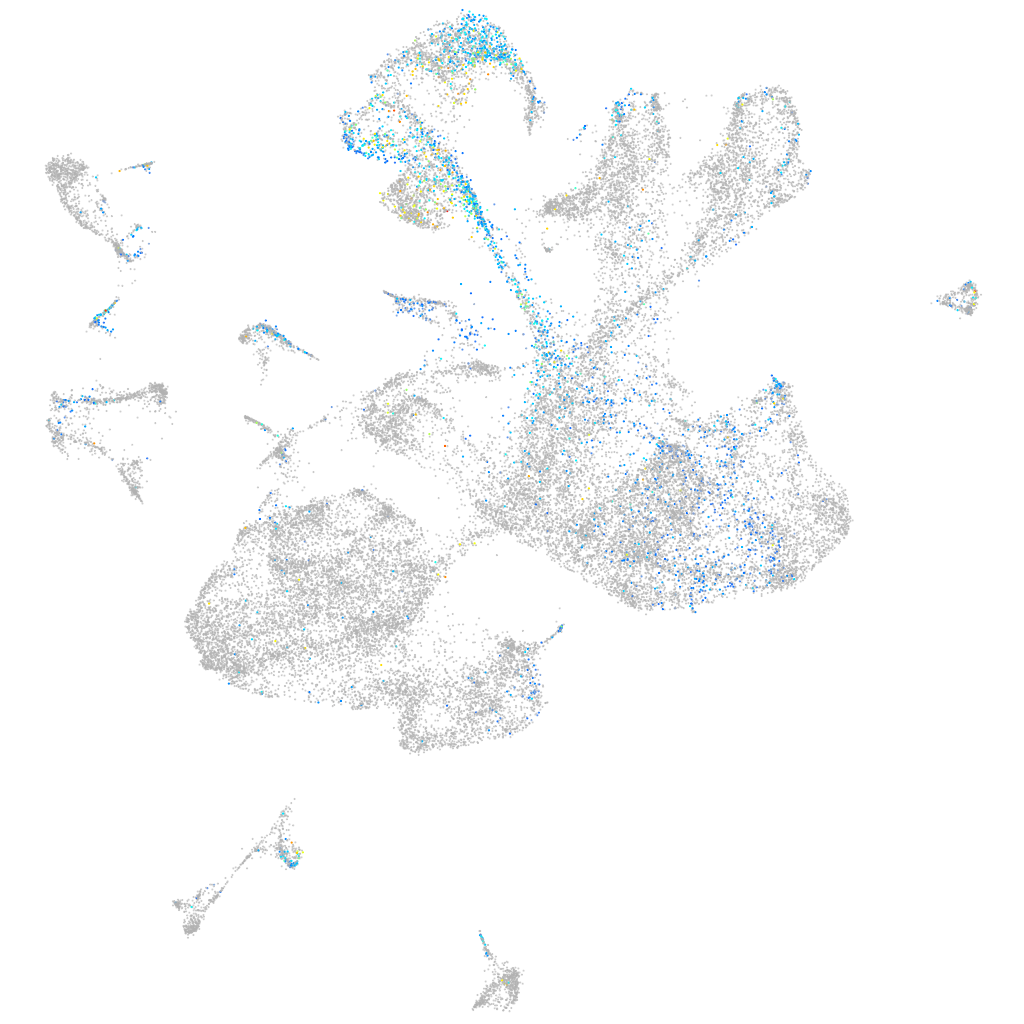
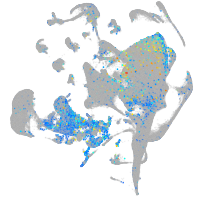
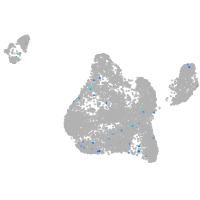
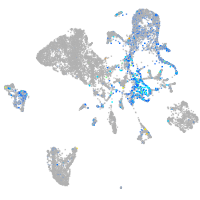
protein tyrosine phosphatase receptor type Fa
ZFIN 
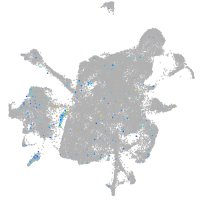
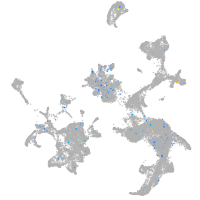
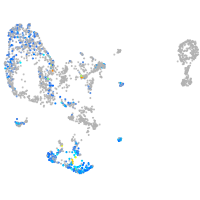
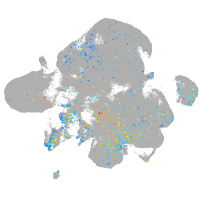
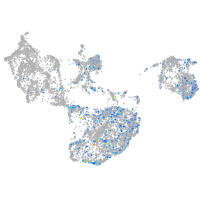
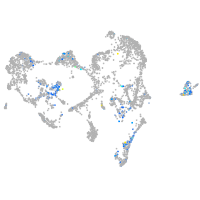
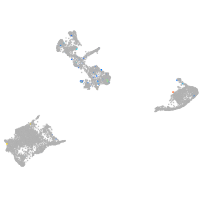
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lhx4 | 0.258 | id1 | -0.129 |
| slit3 | 0.245 | fabp7a | -0.128 |
| isl2a | 0.241 | atp1a1b | -0.118 |
| isl1 | 0.235 | CU467822.1 | -0.117 |
| slc18a3a | 0.222 | zgc:165461 | -0.111 |
| mnx2b | 0.220 | mdkb | -0.109 |
| prph | 0.217 | XLOC-003690 | -0.108 |
| mnx1 | 0.212 | slc1a2b | -0.104 |
| chodl | 0.212 | cd82a | -0.104 |
| cxcr4b | 0.199 | si:ch211-251b21.1 | -0.104 |
| mnx2a | 0.184 | sox3 | -0.103 |
| onecut1 | 0.182 | gpm6bb | -0.102 |
| agrn | 0.181 | mdka | -0.102 |
| l1cama | 0.175 | atp1b4 | -0.100 |
| alcama | 0.174 | msi1 | -0.098 |
| slc5a7a | 0.171 | her15.1 | -0.098 |
| tac1 | 0.171 | notch3 | -0.096 |
| gap43 | 0.168 | qki2 | -0.095 |
| tuba8l3 | 0.166 | CR848047.1 | -0.094 |
| map1b | 0.166 | sparc | -0.093 |
| nrp1a | 0.163 | ptn | -0.087 |
| elavl3 | 0.163 | gas1b | -0.086 |
| ribc1 | 0.162 | slc6a9 | -0.085 |
| gfra3 | 0.161 | cldn5a | -0.084 |
| LOC100537369 | 0.159 | sox2 | -0.084 |
| nrg1 | 0.156 | psat1 | -0.083 |
| islr2 | 0.154 | ppap2d | -0.081 |
| chga | 0.154 | endouc | -0.081 |
| ache | 0.153 | sox19a | -0.080 |
| gng2 | 0.153 | boc | -0.080 |
| ret | 0.153 | CU634008.1 | -0.080 |
| cntn2 | 0.152 | vamp3 | -0.078 |
| inab | 0.151 | si:ch211-286b5.5 | -0.078 |
| LHX3 | 0.149 | pgrmc1 | -0.077 |
| chata | 0.149 | cd63 | -0.077 |