NK2 homeobox 4a
ZFIN 
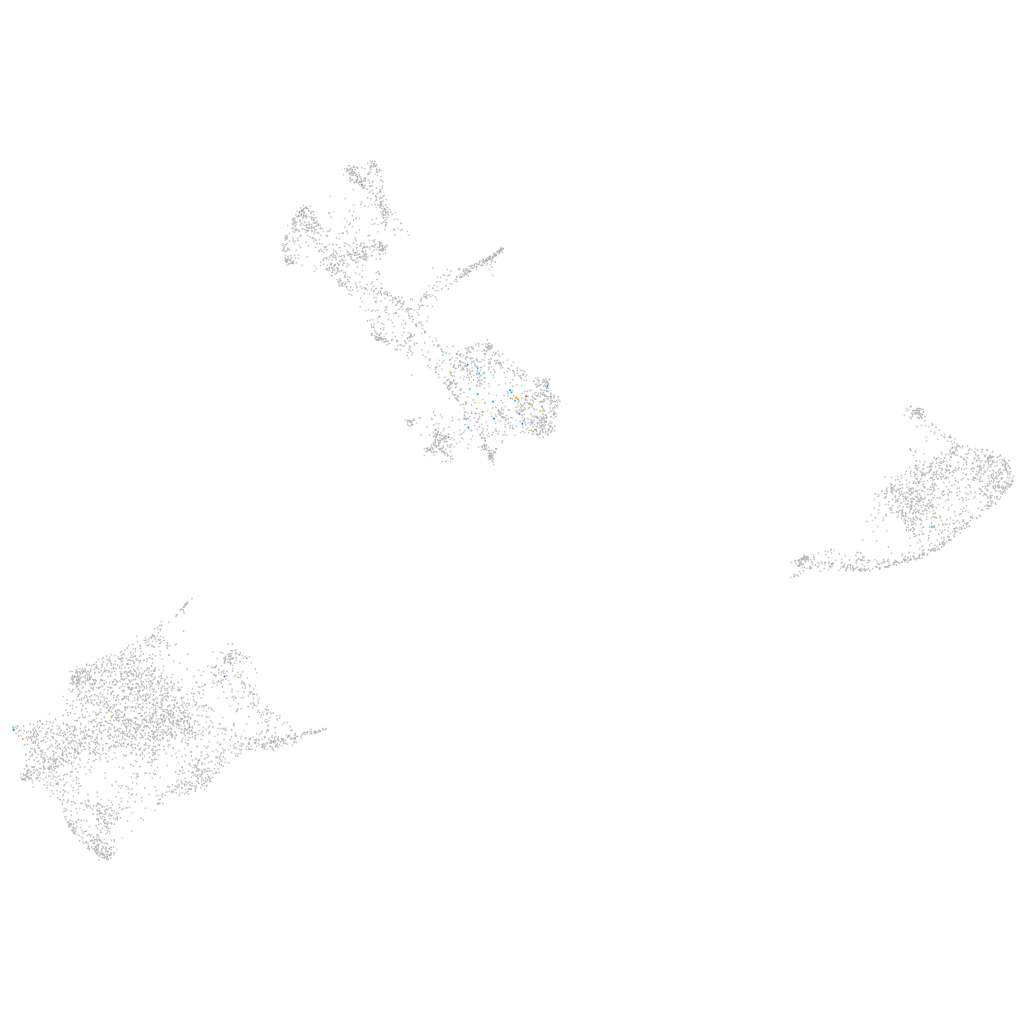
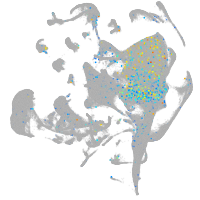
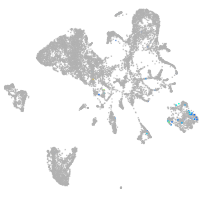
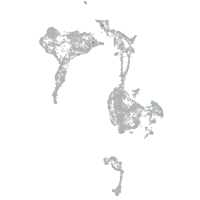
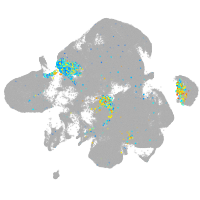
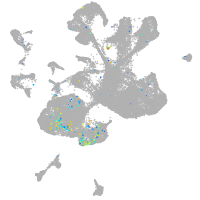
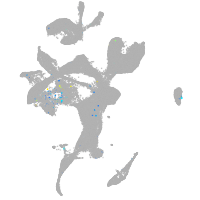
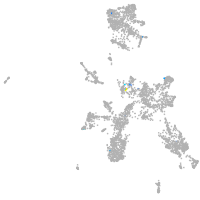
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nkx2.4b | 0.459 | pfn1 | -0.056 |
| CABZ01069436.1 | 0.322 | actb2 | -0.056 |
| sox1b | 0.314 | gmps | -0.046 |
| nox5 | 0.291 | cst14a.2 | -0.046 |
| CABZ01039782.1 | 0.241 | akr1b1 | -0.045 |
| elavl2 | 0.237 | tspan36 | -0.045 |
| dnaaf4 | 0.236 | syngr1a | -0.044 |
| slc6a3 | 0.232 | arhgdia | -0.043 |
| cnga2a | 0.231 | coro1ca | -0.041 |
| LOC103910940 | 0.229 | rab32a | -0.040 |
| ptger1c | 0.229 | zgc:153867 | -0.040 |
| itga3a | 0.226 | rab34b | -0.039 |
| si:ch211-131k2.2 | 0.224 | gpr143 | -0.039 |
| prokr1b | 0.220 | smim29 | -0.039 |
| kif17 | 0.216 | glulb | -0.039 |
| CU462913.1 | 0.208 | bloc1s6 | -0.039 |
| foxd2 | 0.203 | impdh1b | -0.038 |
| LOC108183588 | 0.203 | gart | -0.038 |
| trak1b | 0.202 | krt8 | -0.038 |
| lrp2bp | 0.197 | mdh1aa | -0.037 |
| cops6 | 0.197 | shmt1 | -0.037 |
| ENKD1 | 0.192 | gstp1 | -0.037 |
| kif6 | 0.192 | dera | -0.037 |
| LOC110438203 | 0.191 | slc2a15a | -0.036 |
| CABZ01078606.1 | 0.186 | cox6b2 | -0.036 |
| ptprn2 | 0.184 | vamp3 | -0.035 |
| LOC108191691 | 0.184 | pik3c3 | -0.035 |
| XLOC-028531 | 0.184 | phyhd1 | -0.035 |
| cdk5r2b | 0.183 | rab38 | -0.034 |
| LAMP5 | 0.182 | si:dkey-204f11.64 | -0.034 |
| nkx2.1 | 0.182 | pnp4a | -0.033 |
| raver2 | 0.179 | si:ch211-108c6.2 | -0.033 |
| nmur3 | 0.179 | gch2 | -0.033 |
| tex9 | 0.178 | pts | -0.033 |
| zc2hc1c | 0.176 | pttg1ipb | -0.033 |