nuclear receptor coactivator 3
ZFIN 
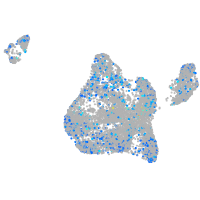
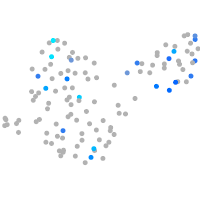
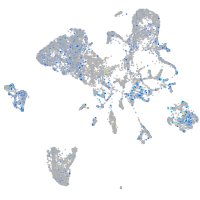
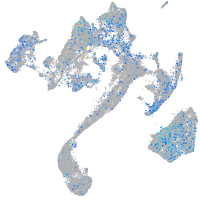
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dhrs3b | 0.089 | atp1a1b | -0.054 |
| hoxb3a | 0.087 | glula | -0.053 |
| hoxc1a | 0.086 | efhd1 | -0.051 |
| hoxb5b | 0.078 | mt2 | -0.051 |
| hoxb6b | 0.073 | fabp7a | -0.050 |
| phox2bb | 0.068 | slc1a2b | -0.048 |
| marcksb | 0.067 | ppap2d | -0.047 |
| nova2 | 0.066 | cox4i2 | -0.046 |
| meis1b | 0.066 | cebpd | -0.046 |
| lin28a | 0.063 | ptgdsb.2 | -0.045 |
| nucks1a | 0.063 | cx43 | -0.045 |
| pbx4 | 0.063 | apoa2 | -0.044 |
| si:ch73-46j18.5 | 0.063 | acbd7 | -0.043 |
| cirbpb | 0.062 | si:ch211-66e2.5 | -0.043 |
| CABZ01075068.1 | 0.060 | slc3a2a | -0.042 |
| hnrnpaba | 0.060 | smox | -0.042 |
| XLOC-016165 | 0.060 | sept8b | -0.041 |
| tubb2b | 0.059 | cd63 | -0.041 |
| h3f3d | 0.058 | mdkb | -0.041 |
| hnrnpa1a | 0.058 | ptn | -0.041 |
| onecut1 | 0.057 | pvalb2 | -0.040 |
| hoxc4a | 0.057 | slc4a4a | -0.040 |
| hoxb1b | 0.057 | mfge8a | -0.039 |
| CR812832.1 | 0.057 | ptgdsb.1 | -0.039 |
| khdrbs1a | 0.057 | cdo1 | -0.039 |
| tmeff1b | 0.057 | eno1b | -0.039 |
| tubb5 | 0.057 | CU467822.1 | -0.039 |
| si:ch211-288g17.3 | 0.056 | zwi | -0.039 |
| ncam1a | 0.056 | apoa1b | -0.038 |
| zgc:100920 | 0.056 | pvalb1 | -0.037 |
| smarce1 | 0.056 | gpr37l1b | -0.036 |
| hnrnpa0l | 0.056 | hspb15 | -0.036 |
| onecut2 | 0.055 | prss59.2 | -0.035 |
| rxrga | 0.055 | hepacama | -0.035 |
| elavl3 | 0.055 | mag | -0.035 |