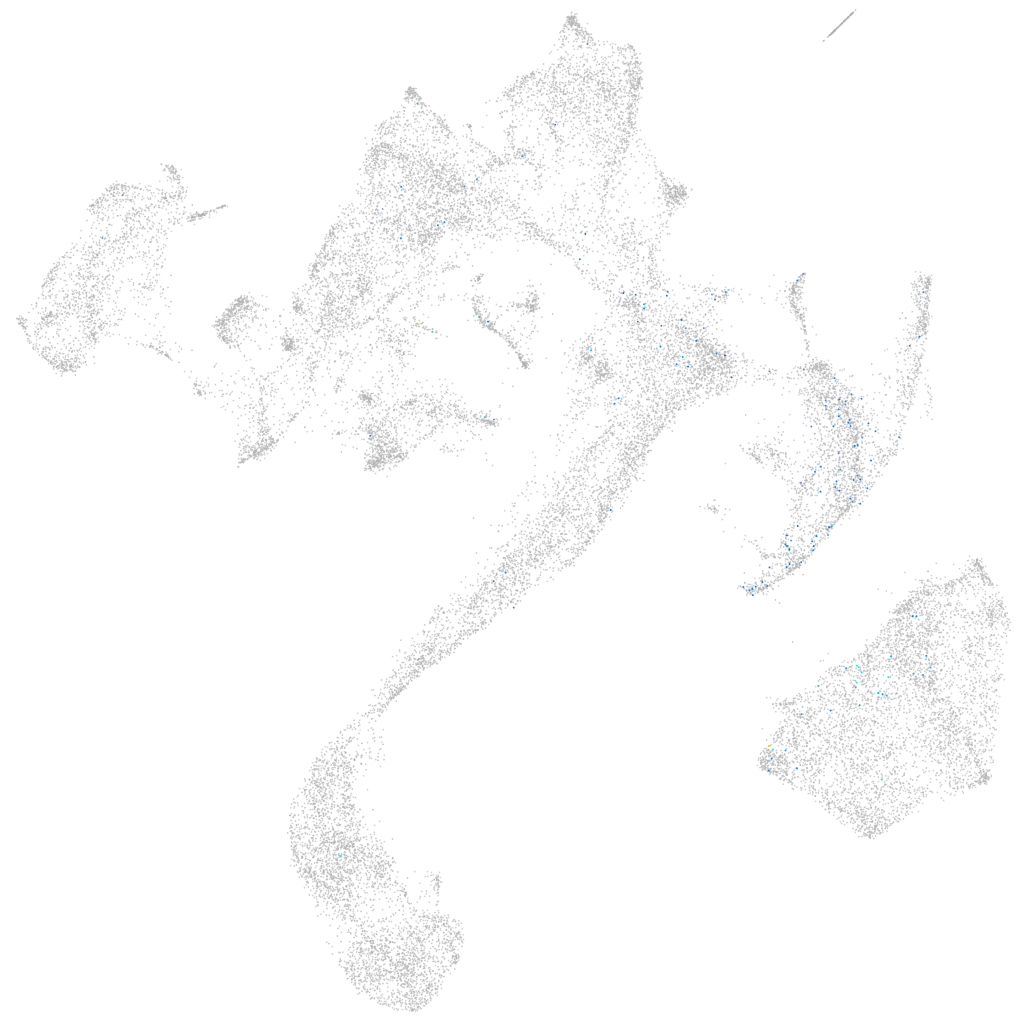
microRNA 196b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia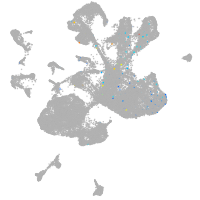 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cd4-2.2 | 0.161 | actc1b | -0.044 |
| BX004816.1 | 0.152 | eef1da | -0.039 |
| CR788249.2 | 0.136 | fabp3 | -0.038 |
| XLOC-007707 | 0.122 | eno1a | -0.038 |
| LOC100537504 | 0.118 | pabpc4 | -0.038 |
| LOC110440044 | 0.110 | atp2a1 | -0.037 |
| LOC110437791 | 0.110 | ckma | -0.037 |
| FAM163B | 0.101 | tmem38a | -0.036 |
| LOC103910432 | 0.099 | ckmb | -0.036 |
| LOC110437710 | 0.099 | ak1 | -0.036 |
| CU660013.1 | 0.098 | mylpfa | -0.035 |
| CU896647.1 | 0.094 | aldoab | -0.035 |
| BX005254.3 | 0.083 | ttn.2 | -0.035 |
| RF00066 | 0.081 | FQ323156.1 | -0.035 |
| aifm3 | 0.080 | gapdh | -0.035 |
| hoxb7a | 0.077 | sparc | -0.034 |
| CR385065.1 | 0.074 | gamt | -0.034 |
| msx1a | 0.074 | desma | -0.034 |
| gucy2g | 0.073 | nme2b.2 | -0.034 |
| hoxc3a | 0.072 | tnnc2 | -0.033 |
| pcdh2ac | 0.071 | klhl41b | -0.033 |
| hoxb10a | 0.070 | neb | -0.033 |
| hoxd10a | 0.070 | pvalb2 | -0.033 |
| her12 | 0.069 | cav3 | -0.033 |
| XLOC-019772 | 0.069 | bhmt | -0.032 |
| mxtx2 | 0.069 | atp5if1b | -0.032 |
| edil3b | 0.069 | pvalb1 | -0.032 |
| itm2cb | 0.068 | acta1b | -0.032 |
| fgf8a | 0.068 | myl1 | -0.032 |
| hes6 | 0.068 | ank1a | -0.032 |
| CR925798.1 | 0.068 | gatm | -0.032 |
| cdkn2a/b | 0.068 | srl | -0.032 |
| LOC103911482 | 0.067 | mdh2 | -0.032 |
| AL929017.1 | 0.065 | acta1a | -0.032 |
| her7 | 0.065 | mybphb | -0.032 |