mesenchyme homeobox 2b
ZFIN 
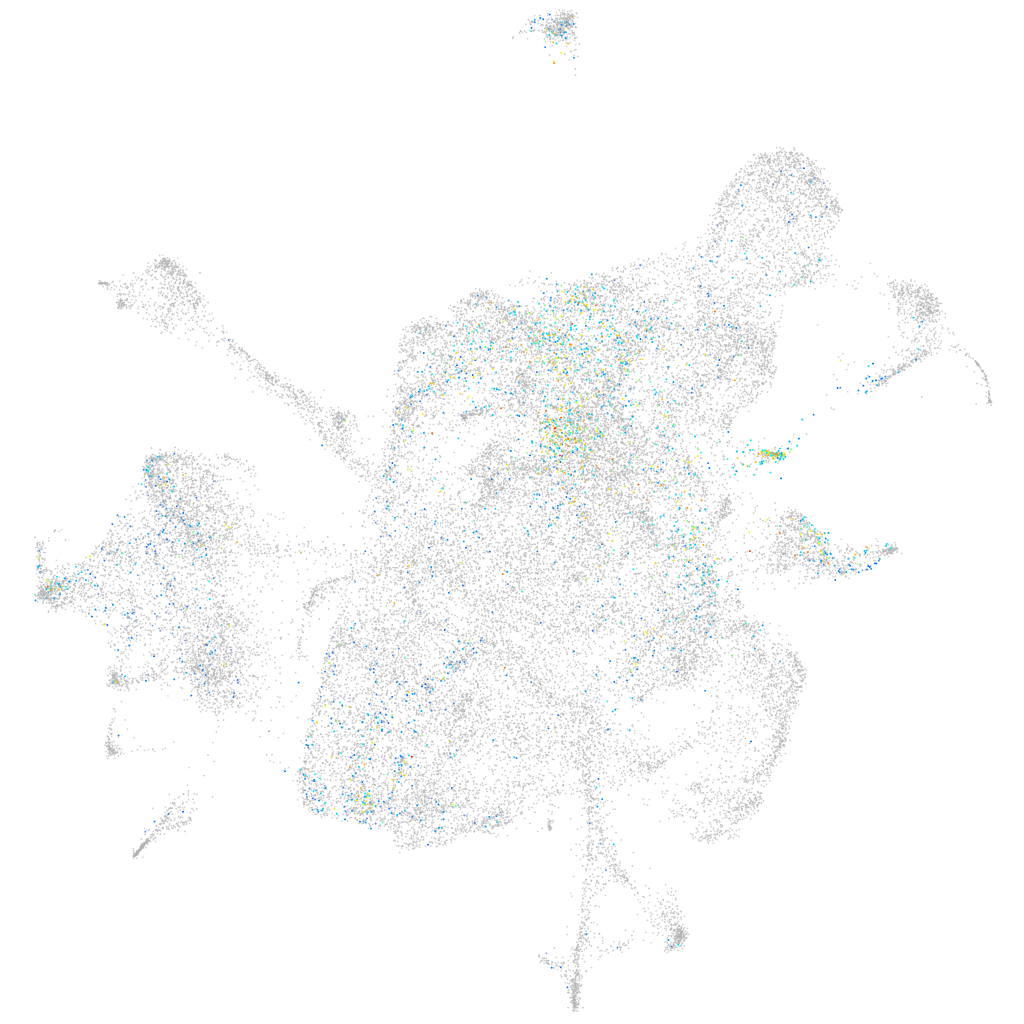
Other cell groups


all cells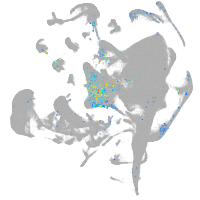 |
blastomeres |
PGCs |
endoderm |
axial mesoderm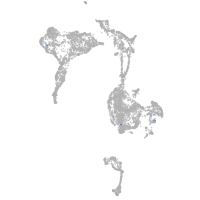 |
muscle 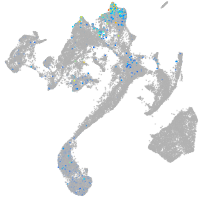 |
mural cells / non-skeletal muscle  |
mesenchyme 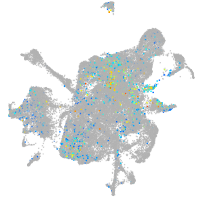 |
hematopoietic / vasculature 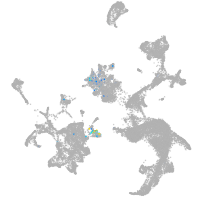 |
pronephros  |
neural |
spinal cord / glia |
eye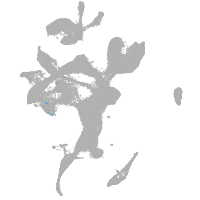 |
taste / olfactory |
otic / lateral line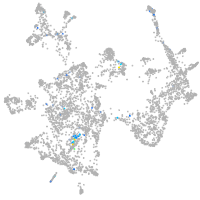 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| shox2 | 0.164 | col11a1a | -0.071 |
| foxf2a | 0.149 | akap12b | -0.062 |
| ecrg4a | 0.146 | col2a1a | -0.061 |
| foxf1 | 0.142 | fabp3 | -0.057 |
| osr2 | 0.134 | lin28a | -0.057 |
| vwde | 0.130 | cdon | -0.056 |
| barx1 | 0.126 | VIT | -0.052 |
| foxq1a | 0.120 | col9a1a | -0.051 |
| foxl1 | 0.116 | ca6 | -0.049 |
| ogna | 0.115 | pitx2 | -0.048 |
| mxra8b | 0.114 | alcama | -0.048 |
| col1a2 | 0.112 | col9a3 | -0.048 |
| gsc | 0.107 | matn4 | -0.048 |
| col5a2a | 0.107 | col11a2 | -0.047 |
| cilp | 0.105 | cthrc1a | -0.047 |
| c1qtnf5 | 0.102 | fstl1a | -0.047 |
| hhip | 0.102 | col9a2 | -0.047 |
| spon2b | 0.102 | hapln1b | -0.047 |
| zgc:162612 | 0.100 | fxyd1 | -0.046 |
| thbs4b | 0.098 | ptges3b | -0.046 |
| twist2 | 0.097 | cnmd | -0.046 |
| col6a2 | 0.097 | tmem88b | -0.046 |
| lhx6 | 0.097 | snrpd1 | -0.045 |
| zgc:162939 | 0.096 | snrpe | -0.045 |
| meox2a | 0.096 | thbs1b | -0.044 |
| hoxd4a | 0.095 | alx4a | -0.044 |
| ntd5 | 0.094 | jam2b | -0.044 |
| col1a1b | 0.094 | abcf1 | -0.044 |
| efnb2b | 0.093 | CABZ01072614.1 | -0.043 |
| col6a1 | 0.093 | epyc | -0.043 |
| LOC108180013 | 0.093 | si:dkey-56m19.5 | -0.043 |
| foxf2b | 0.092 | CABZ01075068.1 | -0.043 |
| efna2a | 0.092 | snrpf | -0.043 |
| meox1 | 0.092 | apex1 | -0.042 |
| ier2b | 0.090 | marcksl1b | -0.042 |