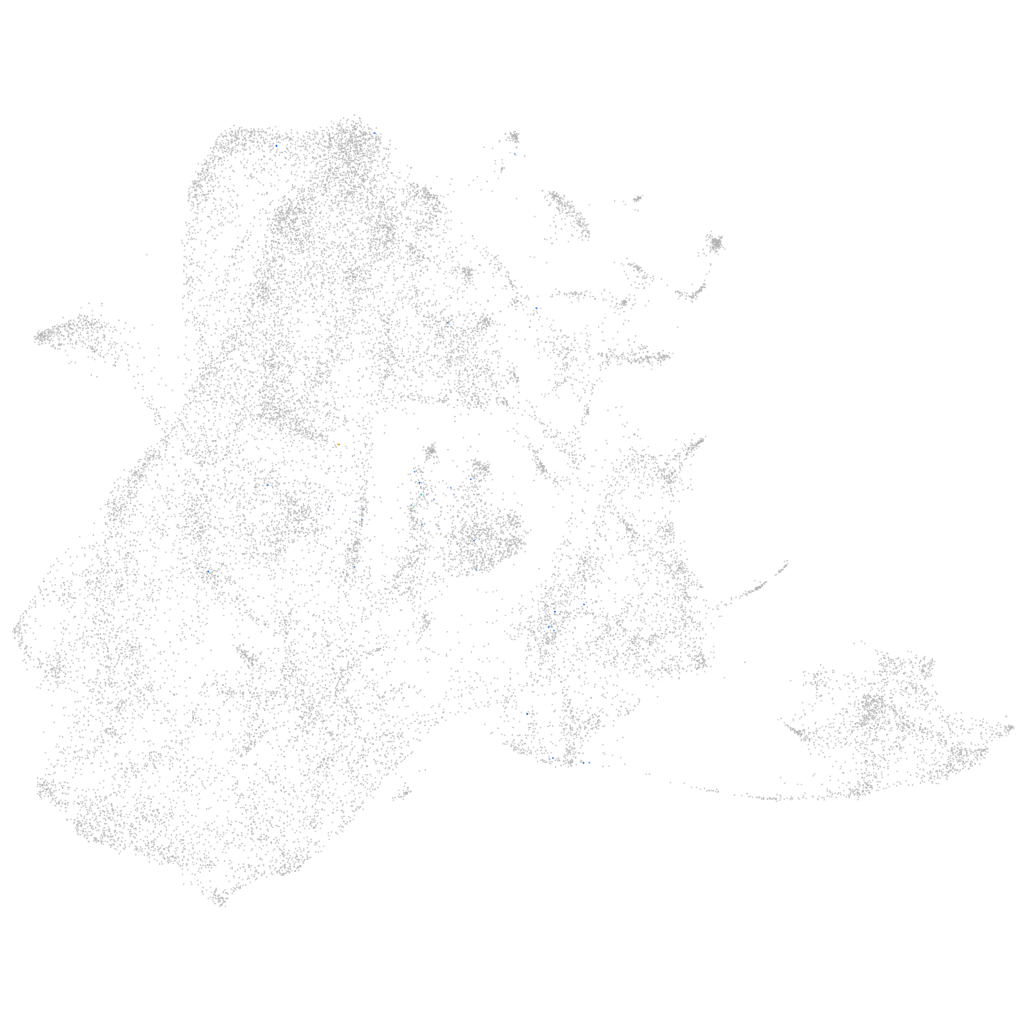
mesenchyme homeobox 2b
ZFIN 
Other cell groups


all cells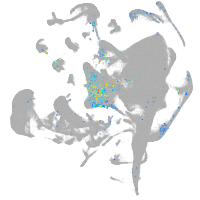 |
blastomeres |
PGCs |
endoderm |
axial mesoderm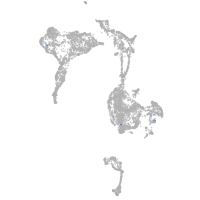 |
muscle 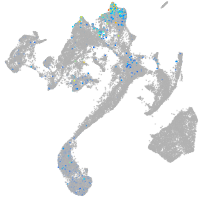 |
mural cells / non-skeletal muscle  |
mesenchyme 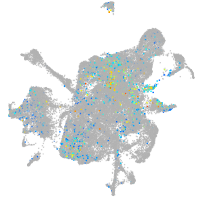 |
hematopoietic / vasculature 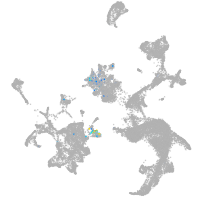 |
pronephros  |
neural |
spinal cord / glia |
eye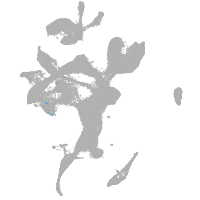 |
taste / olfactory |
otic / lateral line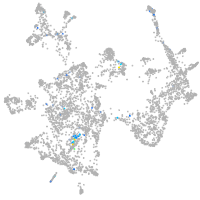 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nkl.4 | 0.308 | cfl1l | -0.037 |
| CR456642.1 | 0.242 | pfn1 | -0.034 |
| tacr2 | 0.212 | krt4 | -0.034 |
| CU929237.1 | 0.210 | tmsb1 | -0.033 |
| gdf6b | 0.207 | actb2 | -0.028 |
| twist1b | 0.196 | cyt1 | -0.027 |
| cd248a | 0.186 | s100a10b | -0.023 |
| BX470224.2 | 0.180 | cldni | -0.022 |
| col6a2 | 0.179 | gsta.1 | -0.022 |
| aspn | 0.167 | arpc3 | -0.021 |
| gsc | 0.161 | myh9a | -0.021 |
| phldb2a | 0.158 | btg2 | -0.020 |
| c1qtnf5 | 0.151 | mapk12a | -0.019 |
| RF00019 | 0.150 | ywhaz | -0.019 |
| mkxa | 0.149 | arhgef5 | -0.019 |
| ssrp1b | 0.145 | hdlbpa | -0.019 |
| pde3a | 0.144 | apoeb | -0.018 |
| igfn1.3 | 0.143 | jupa | -0.018 |
| entpd1 | 0.142 | epcam | -0.018 |
| qsox1 | 0.135 | wu:fb18f06 | -0.018 |
| LOC108180013 | 0.133 | cldn1 | -0.018 |
| LOC103911884 | 0.130 | lgals3b | -0.017 |
| sgcd | 0.126 | ecrg4b | -0.017 |
| antxr1c | 0.124 | tfap2c | -0.017 |
| zgc:162612 | 0.122 | si:rp71-77l1.1 | -0.017 |
| dkk3b | 0.122 | col18a1a | -0.017 |
| rerglb | 0.119 | sh3gl1a | -0.017 |
| SCARF2 | 0.116 | pak2a | -0.016 |
| prelp | 0.116 | tmsb4x | -0.016 |
| FP016205.1 | 0.114 | tp63 | -0.016 |
| LOC110440007 | 0.114 | tax1bp3 | -0.016 |
| mecom | 0.113 | myl12.1 | -0.016 |
| scxa | 0.112 | rpl27 | -0.016 |
| reck | 0.109 | sdr16c5a | -0.016 |
| DGKI | 0.108 | anxa1a | -0.016 |