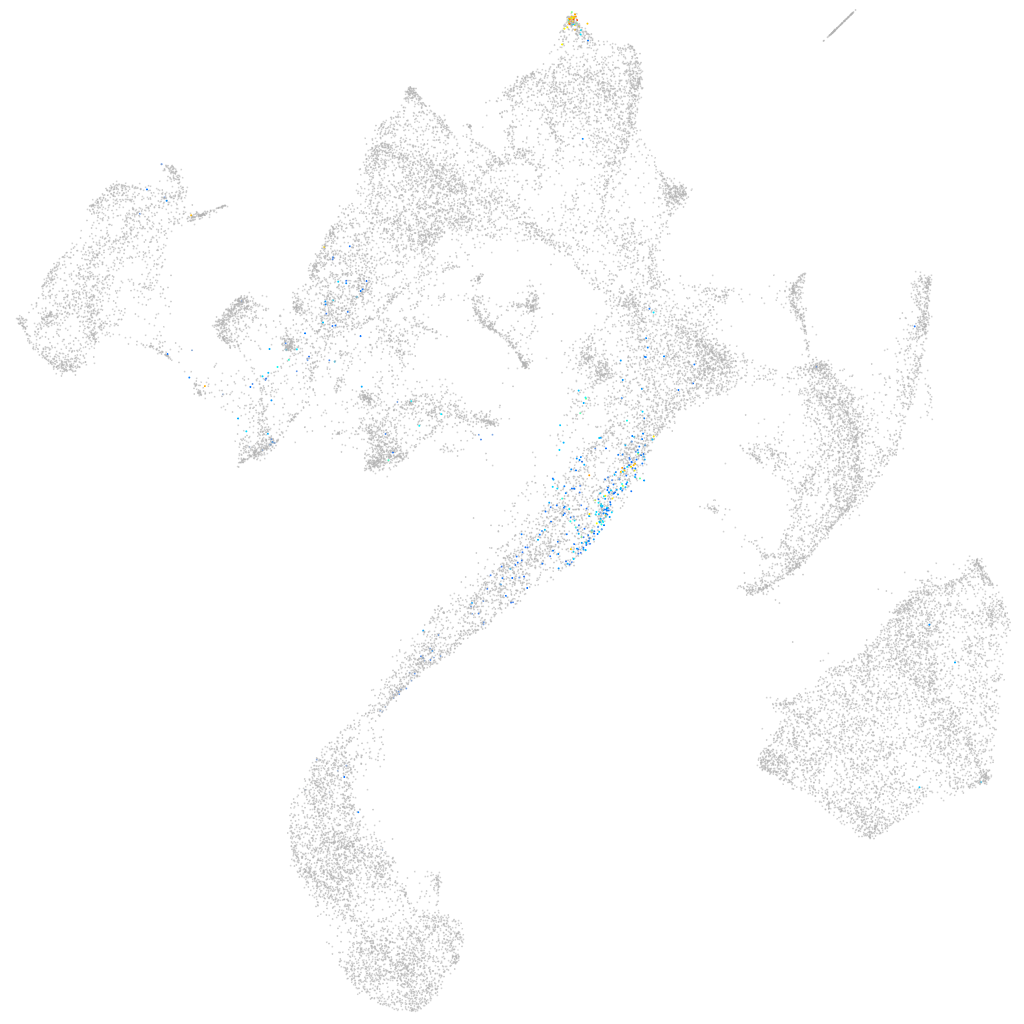
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ecrg4a | 0.257 | ptmab | -0.076 |
| ucmaa | 0.238 | si:ch211-222l21.1 | -0.069 |
| fgf8b | 0.221 | stmn1a | -0.062 |
| r3hdml | 0.216 | pcna | -0.061 |
| prelp | 0.203 | hmgb1b | -0.058 |
| tnmd | 0.196 | nasp | -0.058 |
| prp | 0.176 | tubb2b | -0.058 |
| si:ch211-43f4.1 | 0.173 | cbx5 | -0.057 |
| hlx1 | 0.173 | apoc1 | -0.055 |
| scxa | 0.170 | hmgb2a | -0.054 |
| myog | 0.170 | si:dkey-16p21.8 | -0.054 |
| thrap3a | 0.165 | chaf1a | -0.054 |
| mafaa | 0.159 | rbbp4 | -0.053 |
| mafbb | 0.158 | lbr | -0.052 |
| pcdh10a | 0.156 | anp32b | -0.051 |
| frem1b | 0.155 | myf5 | -0.051 |
| gpc6a | 0.151 | cdx4 | -0.050 |
| jam2a | 0.151 | calm2b | -0.050 |
| sfrp5 | 0.149 | hes6 | -0.050 |
| ptx4 | 0.145 | mcm6 | -0.050 |
| nitr1m | 0.145 | smc1al | -0.050 |
| fam107b | 0.144 | baz1b | -0.048 |
| si:ch1073-268j14.1 | 0.144 | lig1 | -0.048 |
| fbxl22 | 0.137 | dek | -0.048 |
| sh3d19 | 0.134 | rrm1 | -0.048 |
| rbm24a | 0.132 | CABZ01080702.1 | -0.047 |
| si:dkey-17o15.2 | 0.130 | cdca7a | -0.047 |
| tgfbi | 0.129 | hells | -0.047 |
| ripor3 | 0.129 | mki67 | -0.046 |
| aktip | 0.128 | rpa3 | -0.046 |
| zbtb18 | 0.126 | mcm2 | -0.046 |
| si:ch211-159i8.4 | 0.125 | tpx2 | -0.046 |
| cyp1d1 | 0.125 | banf1 | -0.046 |
| rbpjl | 0.124 | apoeb | -0.046 |
| LOC110439930 | 0.123 | mcm5 | -0.046 |