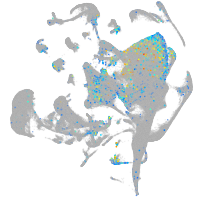
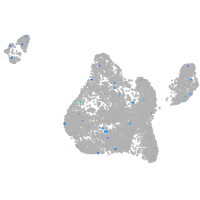
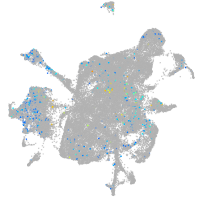
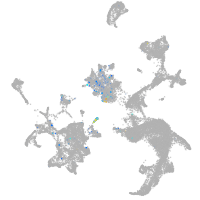
glypican 6a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.136 | aldob | -0.066 |
| snap25a | 0.135 | hspb1 | -0.064 |
| atp6v0cb | 0.134 | apoeb | -0.059 |
| elavl4 | 0.134 | nop58 | -0.058 |
| zgc:65894 | 0.133 | dkc1 | -0.057 |
| scg2b | 0.132 | npm1a | -0.057 |
| ywhag2 | 0.132 | banf1 | -0.056 |
| map1aa | 0.129 | fbl | -0.056 |
| stmn2a | 0.126 | mki67 | -0.056 |
| rtn1b | 0.124 | pou5f3 | -0.056 |
| sv2a | 0.121 | s100a1 | -0.055 |
| stx1b | 0.120 | si:ch211-152c2.3 | -0.055 |
| vamp2 | 0.118 | stm | -0.055 |
| gng3 | 0.117 | apoc1 | -0.053 |
| rnasekb | 0.117 | cfl1l | -0.053 |
| stxbp1a | 0.117 | lig1 | -0.053 |
| atpv0e2 | 0.114 | polr3gla | -0.053 |
| si:dkeyp-75h12.5 | 0.114 | hnrnpa1b | -0.052 |
| sypa | 0.114 | si:dkey-66i24.9 | -0.052 |
| camk2n1a | 0.110 | tpx2 | -0.052 |
| gapdhs | 0.110 | zgc:56699 | -0.052 |
| eno2 | 0.109 | akap12b | -0.051 |
| IGLON5 | 0.108 | arf1 | -0.051 |
| rtn1a | 0.108 | ccnb1 | -0.051 |
| syngr3a | 0.108 | asb11 | -0.050 |
| atp6v1b2 | 0.106 | chaf1a | -0.050 |
| calm1b | 0.106 | epcam | -0.050 |
| gap43 | 0.106 | gar1 | -0.050 |
| gnao1a | 0.106 | zgc:110425 | -0.050 |
| aldocb | 0.105 | anp32b | -0.049 |
| cplx2 | 0.105 | cx43.4 | -0.049 |
| cspg5a | 0.105 | nop56 | -0.049 |
| dnajc5aa | 0.105 | smc4 | -0.049 |
| maptb | 0.105 | vox | -0.049 |
| nrxn1a | 0.105 | ccna2 | -0.048 |