glypican 6a
ZFIN 
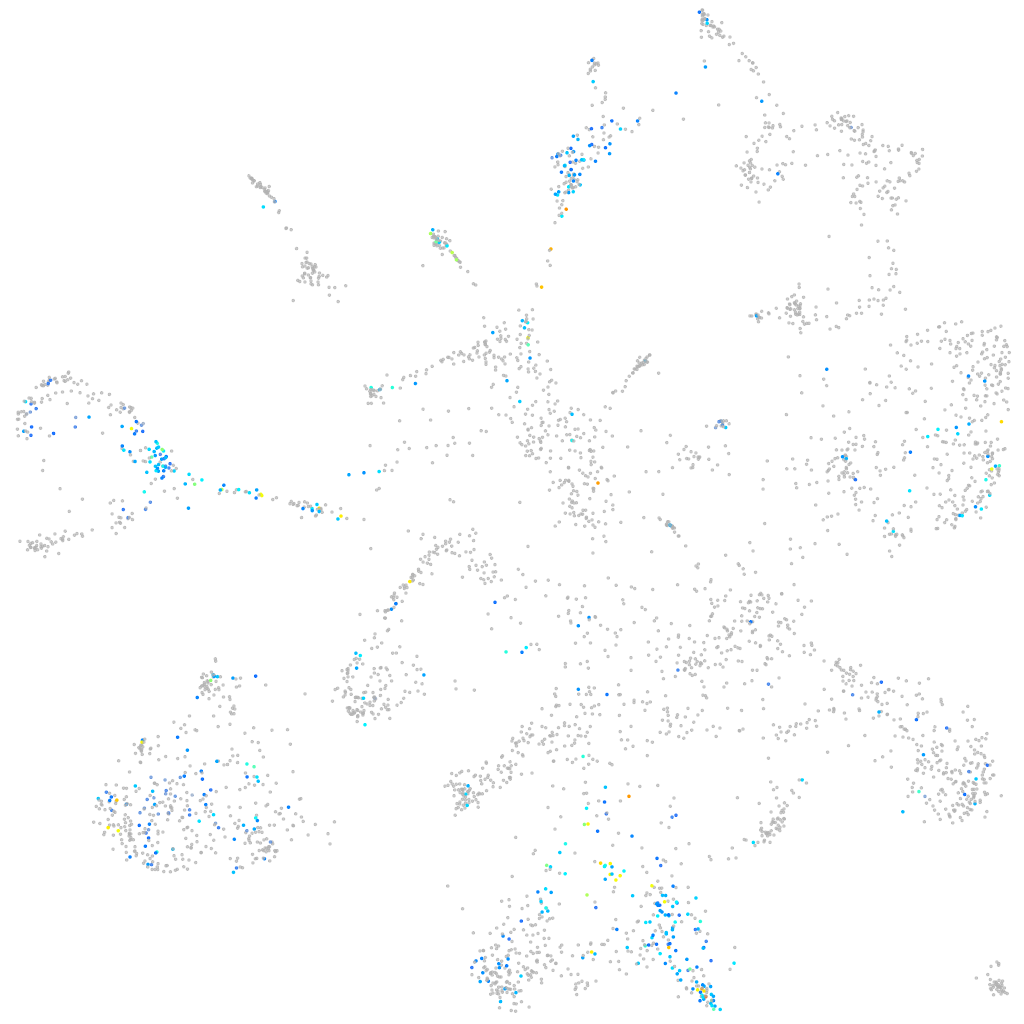
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cald1b | 0.318 | mdka | -0.167 |
| csrp1b | 0.308 | pdgfra | -0.142 |
| acta2 | 0.304 | hmgb2b | -0.104 |
| myh11a | 0.294 | si:dkey-261h17.1 | -0.103 |
| tagln | 0.292 | tmem88b | -0.102 |
| tpm2 | 0.291 | hmga1a | -0.101 |
| mylkb | 0.291 | jam2b | -0.099 |
| cnn1b | 0.289 | fthl27 | -0.098 |
| kcnk18 | 0.289 | marcksl1b | -0.092 |
| actc1a | 0.283 | podxl | -0.092 |
| myl9a | 0.280 | hmgb2a | -0.091 |
| myl6 | 0.272 | nid1b | -0.091 |
| lmod1b | 0.264 | si:ch73-86n18.1 | -0.090 |
| fbxl22 | 0.264 | si:ch1073-291c23.2 | -0.090 |
| tpm1 | 0.263 | mmp16a | -0.088 |
| gucy1a1 | 0.261 | ucp2 | -0.087 |
| cox6b1 | 0.256 | pcna | -0.086 |
| desmb | 0.240 | fgfr2 | -0.085 |
| ckba | 0.239 | stmn1a | -0.084 |
| acta1b | 0.239 | fabp11a | -0.083 |
| acta1a | 0.238 | mcm7 | -0.082 |
| pgm5 | 0.238 | cpn1 | -0.082 |
| mamdc2a | 0.227 | ephb3 | -0.081 |
| tuba8l2 | 0.225 | ets1 | -0.080 |
| smtna | 0.223 | si:ch73-281n10.2 | -0.079 |
| npnt | 0.223 | bambia | -0.079 |
| fsta | 0.221 | ednraa | -0.079 |
| fhl1a | 0.211 | COLEC10 | -0.079 |
| cabp1a | 0.210 | pkp3b | -0.079 |
| vcla | 0.206 | akap12b | -0.078 |
| ppp1cbl | 0.202 | marcksl1a | -0.078 |
| lpp | 0.202 | cdca7b | -0.077 |
| myocd | 0.200 | cebpb | -0.077 |
| atp2b1a | 0.197 | selenoh | -0.074 |
| flna | 0.197 | hmgn2 | -0.074 |