fibroblast growth factor 8 b
ZFIN 
Other cell groups


all cells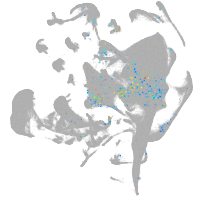 |
blastomeres |
PGCs |
endoderm |
axial mesoderm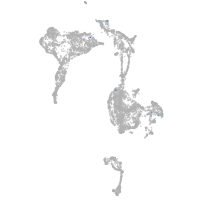 |
muscle  |
mural cells / non-skeletal muscle 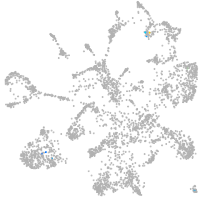 |
mesenchyme 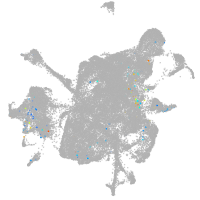 |
hematopoietic / vasculature  |
pronephros 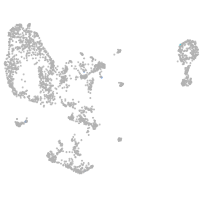 |
neural |
spinal cord / glia |
eye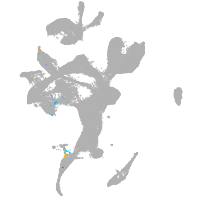 |
taste / olfactory |
otic / lateral line |
epidermis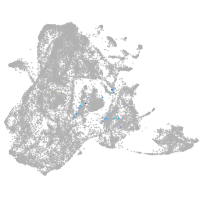 |
periderm |
fin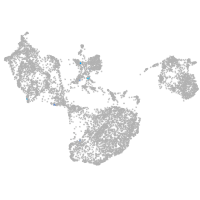 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pax5 | 0.146 | cfl1l | -0.020 |
| slf1 | 0.145 | krt4 | -0.018 |
| en2a | 0.132 | tmsb1 | -0.018 |
| en2b | 0.108 | wu:fb18f06 | -0.018 |
| fgf8a | 0.107 | zgc:153284 | -0.018 |
| en1b | 0.103 | abracl | -0.017 |
| cnpy1 | 0.102 | anxa1a | -0.017 |
| spry2 | 0.091 | cyt1 | -0.017 |
| mlc1 | 0.090 | cyt1l | -0.017 |
| spry4 | 0.088 | myt1a | -0.017 |
| il17rd | 0.087 | myt1b | -0.017 |
| atp1a1b | 0.083 | pfn1 | -0.017 |
| fgf3 | 0.082 | anxa2a | -0.016 |
| slc1a3b | 0.082 | cldne | -0.016 |
| ism1 | 0.080 | dnase1l4.1 | -0.016 |
| pax8 | 0.080 | elavl3 | -0.016 |
| si:ch211-66e2.5 | 0.080 | epcam | -0.016 |
| ppap2d | 0.078 | fgfr3 | -0.016 |
| pax2a | 0.076 | icn | -0.016 |
| si:ch211-43f4.1 | 0.075 | krt5 | -0.016 |
| prp | 0.074 | prr13 | -0.016 |
| mir135b | 0.073 | s100a10b | -0.016 |
| en1a | 0.072 | si:ch211-105c13.3 | -0.016 |
| atp1b4 | 0.071 | si:ch211-195b11.3 | -0.016 |
| mdka | 0.070 | tuba8l2 | -0.016 |
| zcchc7 | 0.069 | agr1 | -0.015 |
| cilp | 0.068 | anxa1c | -0.015 |
| si:ch1073-303k11.2 | 0.066 | arhgef5 | -0.015 |
| si:ch211-159i8.4 | 0.065 | dlx3b | -0.015 |
| vwde | 0.065 | foxq1a | -0.015 |
| prelp | 0.064 | gsnb | -0.015 |
| rgmd | 0.062 | homer2 | -0.015 |
| fgf22 | 0.061 | hrc | -0.015 |
| gpr37l1b | 0.061 | icn2 | -0.015 |
| gpm6bb | 0.060 | lye | -0.015 |




