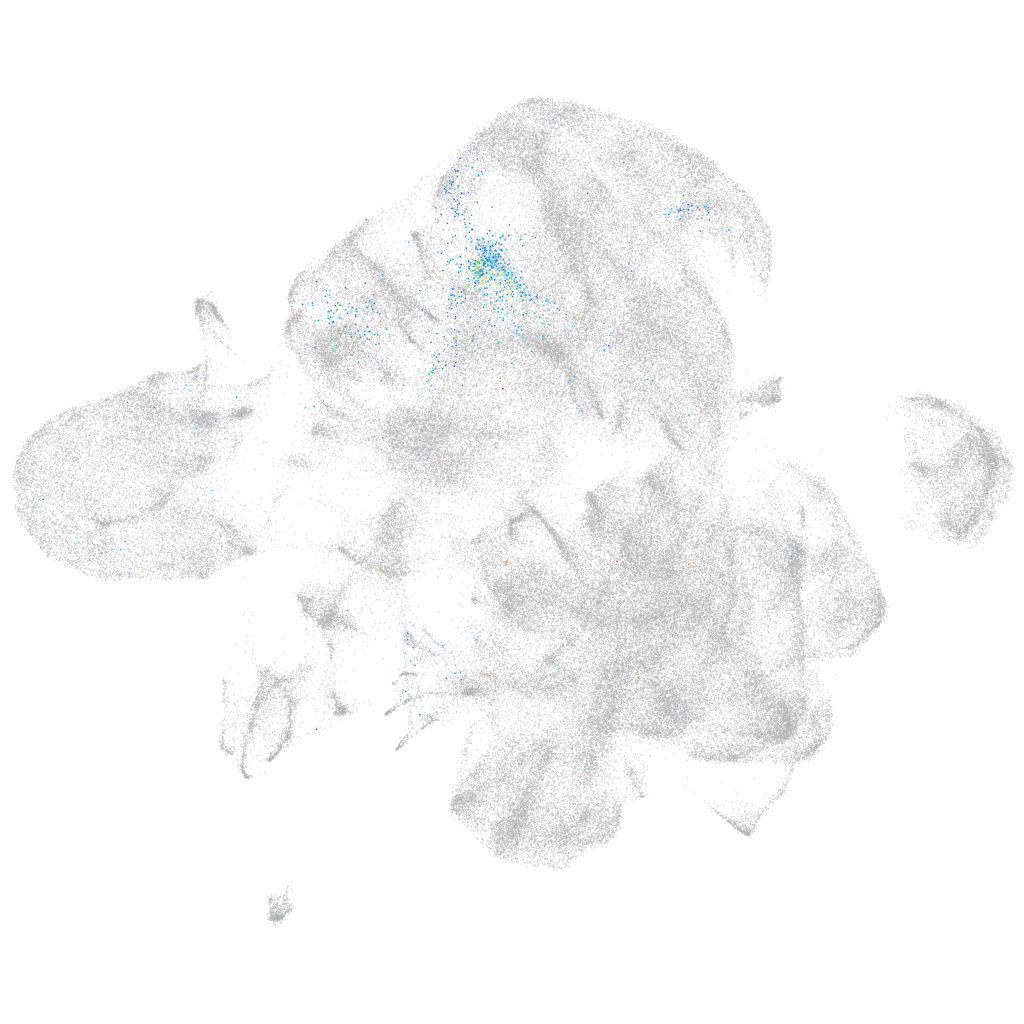
fibroblast growth factor 8 b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slf1 | 0.213 | elavl3 | -0.049 |
| cnpy1 | 0.188 | rtn1a | -0.047 |
| ism1 | 0.184 | gpm6ab | -0.044 |
| pax5 | 0.181 | gng3 | -0.043 |
| il17rd | 0.168 | sncb | -0.041 |
| fgf8a | 0.161 | si:dkey-276j7.1 | -0.041 |
| fgf4 | 0.156 | tubb5 | -0.040 |
| fgf3 | 0.149 | atp6v0cb | -0.039 |
| en2a | 0.149 | ptmaa | -0.039 |
| en1b | 0.141 | rtn1b | -0.039 |
| zcchc7 | 0.128 | stmn1b | -0.039 |
| en2b | 0.126 | hmgb3a | -0.038 |
| en1a | 0.113 | zc4h2 | -0.038 |
| spry2 | 0.104 | elavl4 | -0.037 |
| RNF224 | 0.103 | vamp2 | -0.037 |
| vegfc | 0.102 | fez1 | -0.036 |
| si:ch211-66e2.5 | 0.101 | h2afx1 | -0.036 |
| spry4 | 0.101 | rnasekb | -0.036 |
| gja8a | 0.100 | calm1a | -0.035 |
| atp1a1b | 0.098 | stmn2a | -0.035 |
| pax8 | 0.098 | ywhag2 | -0.035 |
| si:ch211-67e16.11 | 0.098 | stx1b | -0.035 |
| sdc4 | 0.097 | stxbp1a | -0.035 |
| bgnb | 0.094 | mllt11 | -0.034 |
| mir135b | 0.093 | si:dkeyp-75h12.5 | -0.034 |
| olfml2bb | 0.089 | myt1b | -0.033 |
| hmga2 | 0.087 | gap43 | -0.033 |
| nkx2.9 | 0.085 | tuba1c | -0.033 |
| mlc1 | 0.085 | gng2 | -0.033 |
| pax2a | 0.084 | atp6v1e1b | -0.033 |
| mdka | 0.084 | zgc:65894 | -0.033 |
| atp1b4 | 0.082 | myt1a | -0.033 |
| apcdd1l | 0.079 | nsg2 | -0.033 |
| slc1a3b | 0.079 | snap25a | -0.032 |
| afap1l2 | 0.077 | gpm6aa | -0.032 |