homeobox and leucine zipper encoding a
ZFIN 
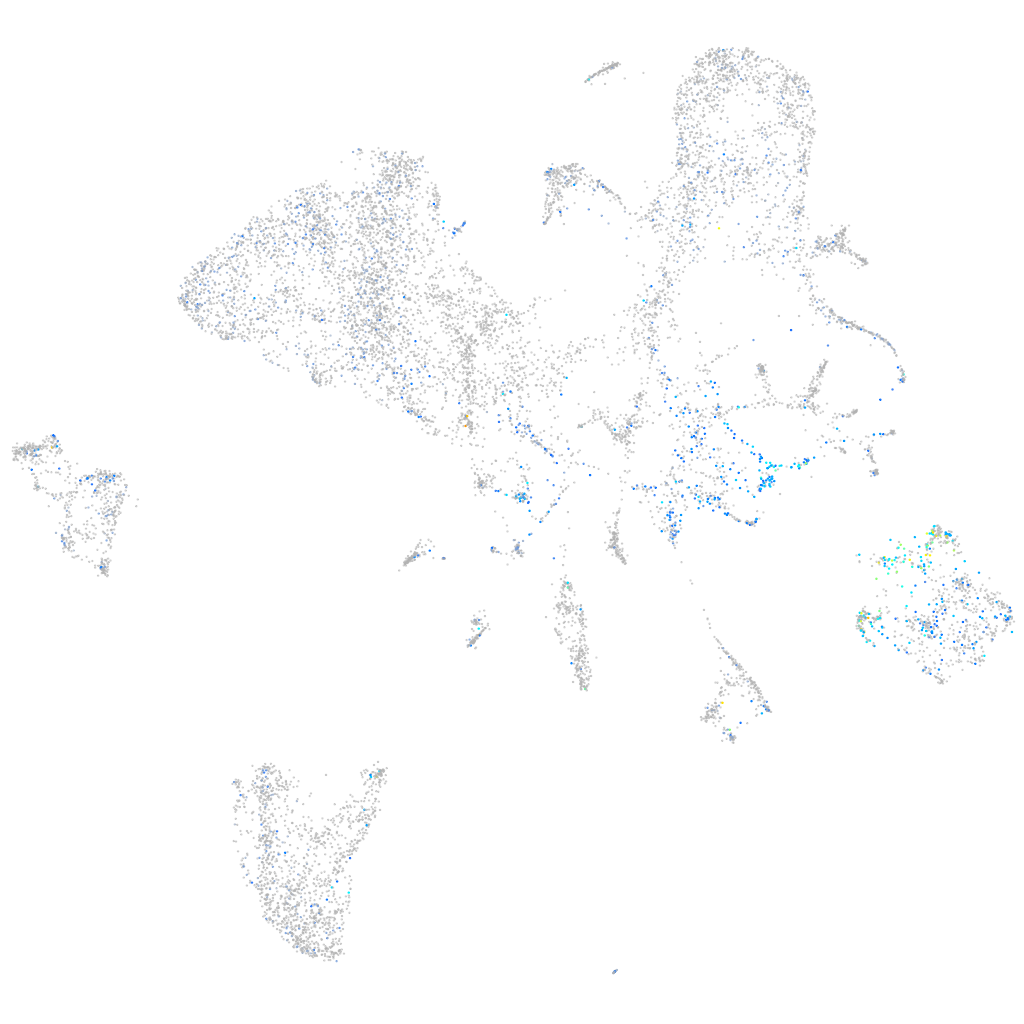
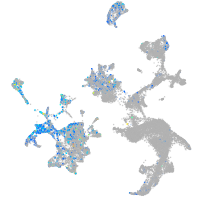
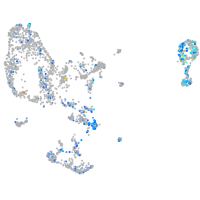
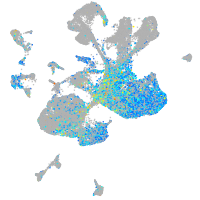
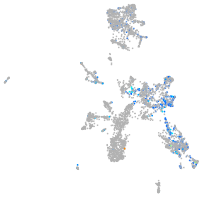
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apela | 0.306 | gapdh | -0.185 |
| pltp | 0.302 | ahcy | -0.184 |
| cdx4 | 0.298 | rpl37 | -0.180 |
| hoxc8a | 0.283 | sod1 | -0.168 |
| cx43.4 | 0.279 | eno3 | -0.167 |
| hspb1 | 0.278 | rps10 | -0.167 |
| cdh6 | 0.273 | gamt | -0.160 |
| BX005254.3 | 0.265 | nme2b.1 | -0.159 |
| qkia | 0.260 | aldob | -0.157 |
| hoxb7a | 0.256 | zgc:114188 | -0.150 |
| fgfrl1b | 0.252 | rps17 | -0.148 |
| crtap | 0.252 | gatm | -0.148 |
| znfl2a | 0.252 | atp5if1b | -0.148 |
| ndnf | 0.251 | atp5l | -0.145 |
| sept15 | 0.249 | tpi1b | -0.144 |
| hoxc6b | 0.248 | eef1da | -0.141 |
| akap12b | 0.248 | suclg1 | -0.139 |
| acin1a | 0.246 | gstp1 | -0.139 |
| marcksb | 0.241 | zgc:158463 | -0.138 |
| hapln1b | 0.241 | gpx4a | -0.136 |
| serpinh1b | 0.240 | lgals2b | -0.134 |
| emid1 | 0.237 | prdx2 | -0.134 |
| si:ch73-281n10.2 | 0.237 | fbp1b | -0.134 |
| hoxc3a | 0.237 | dap | -0.132 |
| prickle1a | 0.236 | nupr1b | -0.132 |
| CABZ01075068.1 | 0.235 | pklr | -0.131 |
| tgif1 | 0.235 | apoc2 | -0.131 |
| nucks1a | 0.234 | apoa4b.1 | -0.130 |
| fgf10b | 0.233 | glud1b | -0.127 |
| hmga1a | 0.233 | zgc:92744 | -0.127 |
| oc90 | 0.231 | apoa1b | -0.127 |
| id3 | 0.229 | bhmt | -0.126 |
| hnrnpaba | 0.229 | BX908782.3 | -0.124 |
| hmgn6 | 0.229 | fabp2 | -0.123 |
| tpm4a | 0.227 | atp5mc1 | -0.123 |