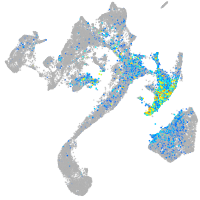
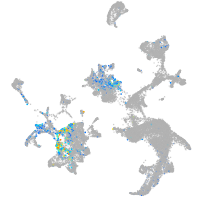
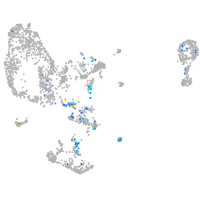
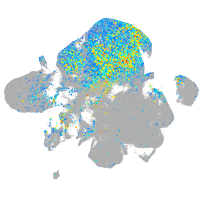
hairy-related 12
ZFIN 
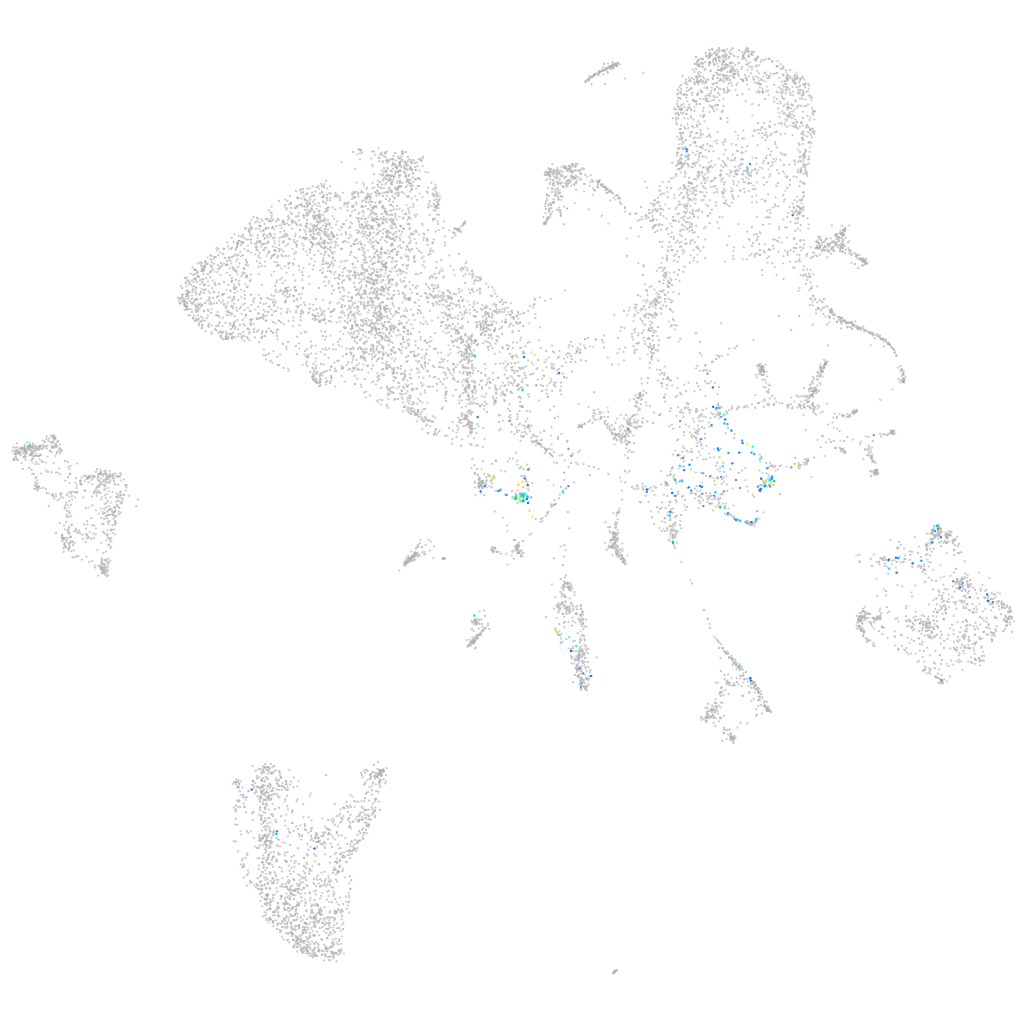
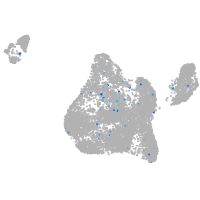
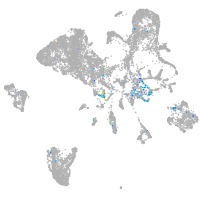
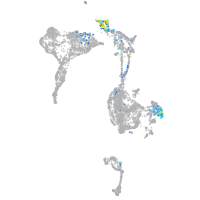
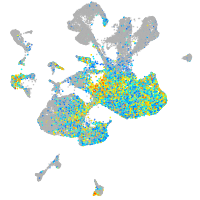
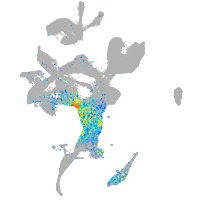
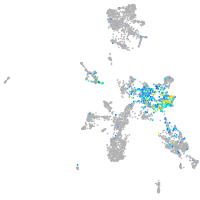
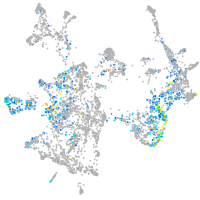
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-003689 | 0.374 | gapdh | -0.126 |
| her4.1 | 0.369 | sod1 | -0.125 |
| XLOC-003690 | 0.365 | eno3 | -0.124 |
| si:ch73-21g5.7 | 0.343 | eef1da | -0.119 |
| XLOC-003692 | 0.339 | aldob | -0.115 |
| abhd6a | 0.327 | atp5if1b | -0.111 |
| her4.2 | 0.314 | BX908782.3 | -0.110 |
| her4.4 | 0.302 | glud1b | -0.108 |
| LOC798783 | 0.298 | nupr1b | -0.108 |
| her15.1 | 0.279 | cebpd | -0.104 |
| mir219-3 | 0.273 | pklr | -0.103 |
| her2 | 0.261 | ahcy | -0.102 |
| CABZ01075068.1 | 0.249 | gstt1a | -0.102 |
| mdka | 0.249 | sod2 | -0.101 |
| cldn5a | 0.249 | cx32.3 | -0.100 |
| pax3a | 0.235 | scp2a | -0.099 |
| fstl1a | 0.232 | gamt | -0.098 |
| ndnf | 0.232 | ckba | -0.097 |
| gdf6b | 0.225 | gstp1 | -0.096 |
| pcdh19 | 0.225 | mat1a | -0.096 |
| sall4 | 0.224 | pgk1 | -0.096 |
| gfap | 0.219 | lgals2b | -0.095 |
| lin28a | 0.216 | ugt1a7 | -0.093 |
| fgfrl1b | 0.211 | fbp1b | -0.093 |
| plp1a | 0.209 | abat | -0.093 |
| msi1 | 0.209 | prdx6 | -0.093 |
| col8a1a | 0.207 | aldh6a1 | -0.093 |
| si:ch211-1e14.1 | 0.206 | sdr16c5b | -0.091 |
| CU634008.1 | 0.204 | gstk1 | -0.091 |
| gas1b | 0.199 | nipsnap3a | -0.088 |
| emid1 | 0.194 | cox8b | -0.088 |
| fbn2b | 0.188 | rmdn1 | -0.088 |
| ap1s2 | 0.188 | agxtb | -0.088 |
| notch1b | 0.187 | gcshb | -0.088 |
| zgc:165461 | 0.186 | sult2st2 | -0.087 |