G protein-coupled receptor 37 like 1a
ZFIN 
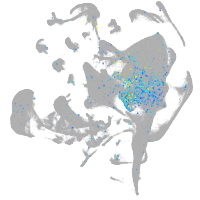
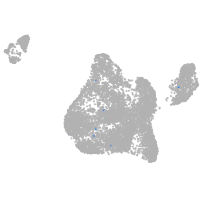
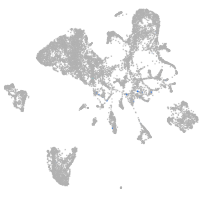
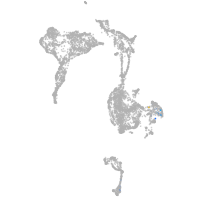
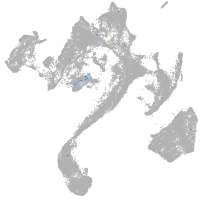
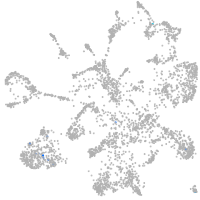
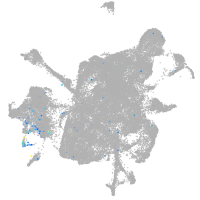
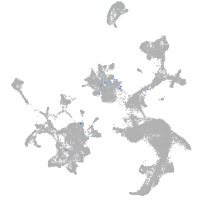
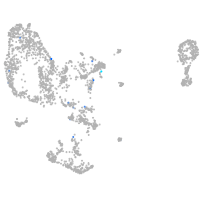
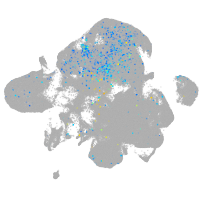
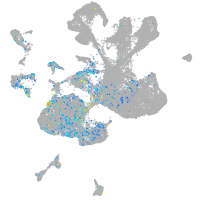
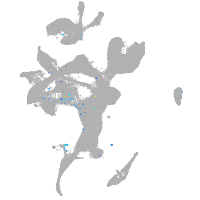
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fabp7a | 0.205 | ptmab | -0.129 |
| si:dkey-183j2.10 | 0.200 | elavl3 | -0.125 |
| CR848047.1 | 0.192 | h3f3d | -0.123 |
| qki2 | 0.176 | si:ch211-222l21.1 | -0.115 |
| CU467822.1 | 0.173 | tubb2b | -0.110 |
| cd82a | 0.173 | hoxb9a | -0.109 |
| nfia | 0.173 | hsp90ab1 | -0.108 |
| gpm6bb | 0.172 | h3f3a | -0.107 |
| metrnla | 0.172 | ptmaa | -0.102 |
| slc1a3b | 0.166 | hoxc3a | -0.096 |
| si:ch211-66e2.5 | 0.162 | zc4h2 | -0.095 |
| notch3 | 0.155 | myt1b | -0.094 |
| slc1a2b | 0.155 | stxbp1a | -0.094 |
| tnc | 0.155 | sumo3a | -0.092 |
| slit2 | 0.154 | hnrnpaba | -0.090 |
| atp1a1b | 0.154 | gng3 | -0.089 |
| fgfr2 | 0.152 | gng2 | -0.089 |
| atp1b1a | 0.150 | si:ch73-386h18.1 | -0.089 |
| ptn | 0.150 | hnrnpa0l | -0.088 |
| atp1b4 | 0.149 | stx1b | -0.088 |
| ntn1b | 0.149 | vamp2 | -0.087 |
| CU634008.1 | 0.147 | sncb | -0.087 |
| igfbp7 | 0.142 | elavl4 | -0.087 |
| podxl | 0.141 | LOC100537384 | -0.086 |
| s100b | 0.141 | snap25a | -0.086 |
| canx | 0.139 | myt1a | -0.085 |
| rpz5 | 0.138 | ube2e1 | -0.084 |
| cnn2 | 0.138 | onecut1 | -0.083 |
| si:ch1073-303k11.2 | 0.136 | stmn1a | -0.083 |
| fgfr3 | 0.134 | hoxa9a | -0.083 |
| gpr37l1b | 0.133 | fthl27 | -0.082 |
| myo18aa | 0.133 | onecut2 | -0.081 |
| cspg5a | 0.132 | nsg2 | -0.081 |
| ptprfb | 0.130 | hoxd3a | -0.081 |
| hepacama | 0.130 | jagn1a | -0.081 |