"finTRIM family, member 83"
ZFIN 
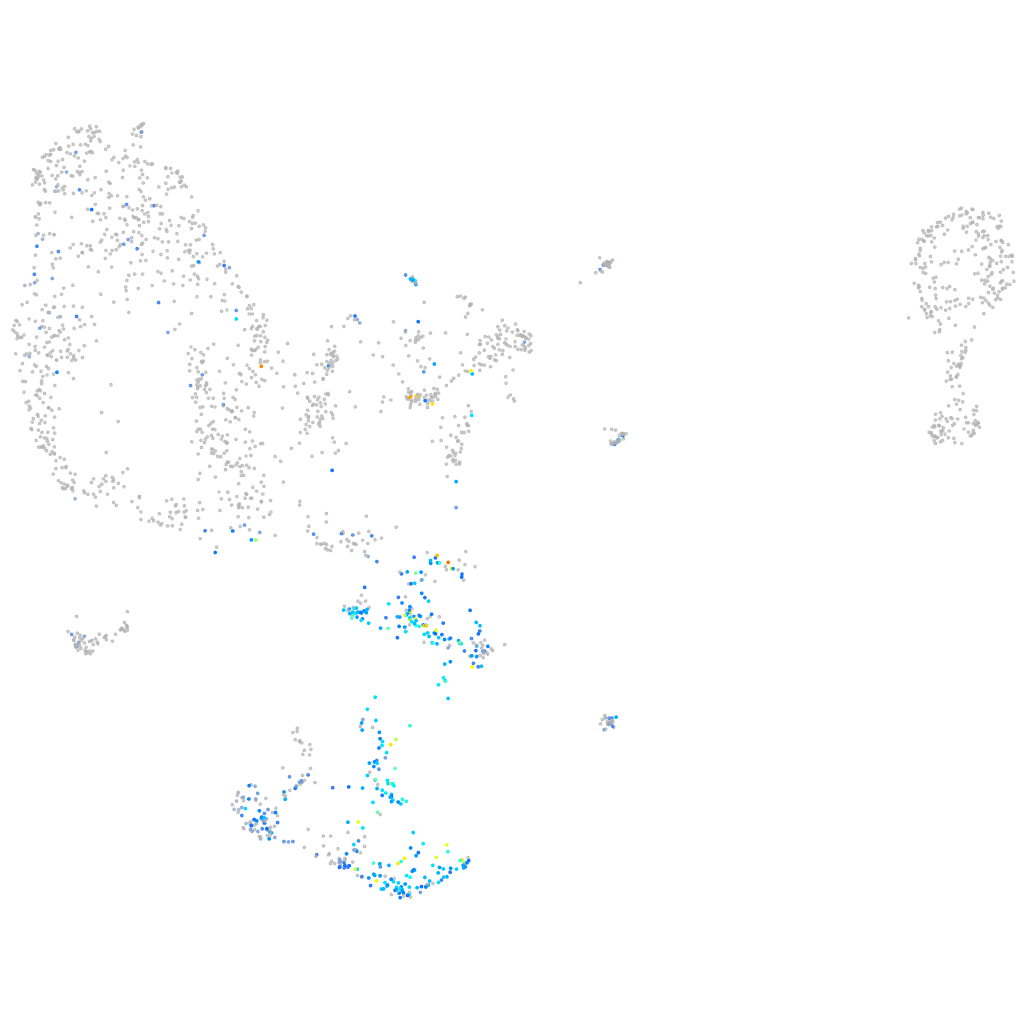
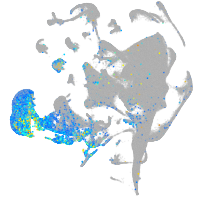
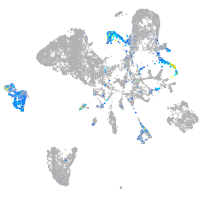
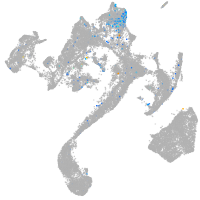
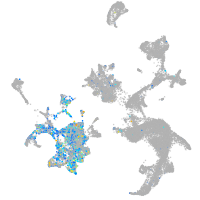
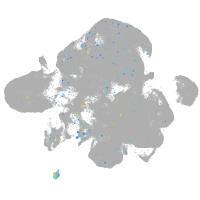
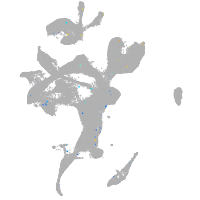
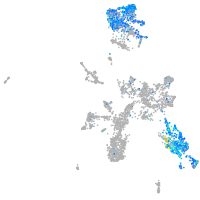
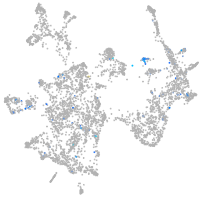
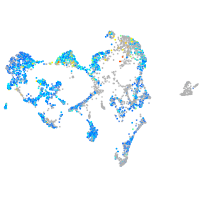
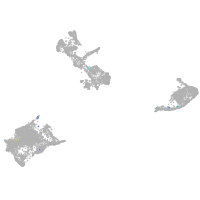
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rnf183 | 0.503 | grhprb | -0.313 |
| ftr82 | 0.489 | dab2 | -0.285 |
| cldnh | 0.466 | pdzk1ip1 | -0.280 |
| phlda2 | 0.444 | hao1 | -0.277 |
| degs2 | 0.438 | fbp1b | -0.275 |
| cldnc | 0.437 | gstr | -0.275 |
| atp1a1a.3 | 0.430 | upb1 | -0.275 |
| si:dkey-36i7.3 | 0.417 | gstt1a | -0.273 |
| cldnb | 0.416 | pklr | -0.273 |
| cldne | 0.410 | si:dkey-194e6.1 | -0.269 |
| mal2 | 0.409 | pnp6 | -0.266 |
| bsnd | 0.407 | agxta | -0.266 |
| hoxc11b | 0.406 | pdzk1 | -0.264 |
| tfap2a | 0.396 | cubn | -0.262 |
| hoxb8a | 0.390 | si:dkey-28n18.9 | -0.261 |
| soul2 | 0.389 | gamt | -0.260 |
| gdpd3a | 0.387 | zgc:136493 | -0.259 |
| txnipa | 0.387 | ahcy | -0.258 |
| si:ch73-359m17.9 | 0.383 | aldob | -0.257 |
| zgc:158423 | 0.380 | si:dkey-33i11.9 | -0.256 |
| hoxc13b | 0.379 | slc26a1 | -0.255 |
| trim35-12 | 0.372 | ftcd | -0.253 |
| etf1a | 0.372 | gnpda1 | -0.251 |
| agr2 | 0.371 | hoga1 | -0.250 |
| tfap2b | 0.368 | alpl | -0.247 |
| mctp2b | 0.366 | dpydb | -0.247 |
| ppp1r1b | 0.366 | nit2 | -0.237 |
| her6 | 0.364 | dpys | -0.237 |
| flvcr1 | 0.363 | ahcyl1 | -0.232 |
| ckmt1 | 0.356 | lrp2a | -0.231 |
| elf3 | 0.352 | adka | -0.231 |
| vcana | 0.351 | lgmn | -0.225 |
| clcnk | 0.351 | suclg2 | -0.225 |
| oclnb | 0.349 | gpt2l | -0.224 |
| tmem72 | 0.348 | cx32.3 | -0.222 |