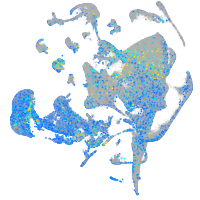
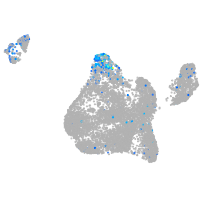
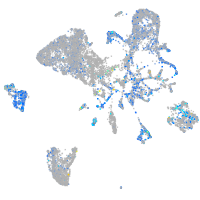
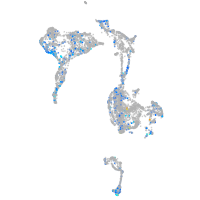
FERM domain containing 4Ba
ZFIN 
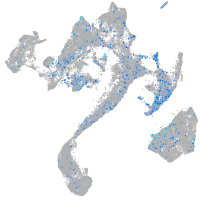
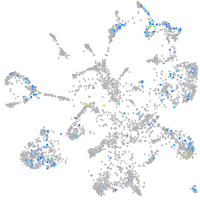
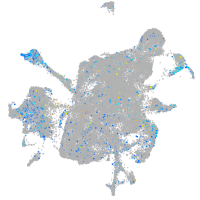
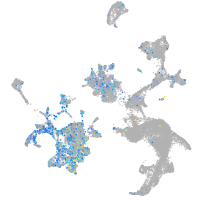
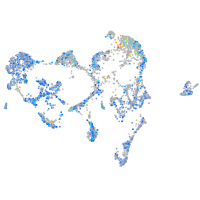
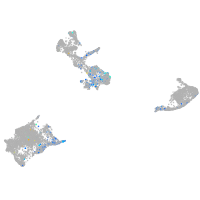
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-164f24.2 | 0.156 | fibina | -0.061 |
| gyg1b | 0.154 | pmp22a | -0.060 |
| wnt9b | 0.140 | matn4 | -0.060 |
| tkta | 0.131 | cdh11 | -0.060 |
| fgl2a | 0.116 | si:ch211-286o17.1 | -0.054 |
| gbe1a | 0.116 | tgfbi | -0.053 |
| samd10a | 0.116 | cd248b | -0.052 |
| hoxa13b | 0.113 | tnc | -0.049 |
| emilin3a | 0.112 | six1a | -0.048 |
| lrrc58a | 0.109 | col11a1b | -0.042 |
| scinla | 0.109 | hmga2 | -0.042 |
| clic2 | 0.107 | foxd1 | -0.041 |
| mgarp | 0.102 | col5a1 | -0.039 |
| cldn5a | 0.099 | col12a1a | -0.039 |
| tubb6 | 0.099 | cthrc1a | -0.037 |
| fam49a | 0.094 | otos | -0.035 |
| timp4.1 | 0.092 | six4a | -0.035 |
| gpc1a | 0.092 | ebf1a | -0.034 |
| tfap2b | 0.091 | ccdc3b | -0.033 |
| alcamb | 0.090 | tuba8l3 | -0.033 |
| cadm4 | 0.088 | col1a1a | -0.033 |
| ccka | 0.087 | CU929237.1 | -0.033 |
| lmx1ba | 0.085 | inka1a | -0.032 |
| qdpra | 0.085 | six2a | -0.032 |
| cyp1b1 | 0.083 | mfap2 | -0.032 |
| tbx5a | 0.083 | si:ch73-86n18.1 | -0.032 |
| pitx1 | 0.082 | cnmd | -0.032 |
| ppp1r1c | 0.082 | ifitm1 | -0.032 |
| pitx2 | 0.081 | foxd2 | -0.031 |
| slc3a2a | 0.078 | foxf2a | -0.031 |
| cotl1 | 0.076 | eya1 | -0.031 |
| col8a2 | 0.074 | snai1a | -0.030 |
| hoxa11b | 0.073 | ednraa | -0.030 |
| col4a3 | 0.072 | serpinf1 | -0.030 |
| f13a1b | 0.072 | hapln1a | -0.030 |