cholecystokinin a
ZFIN 
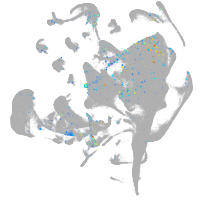
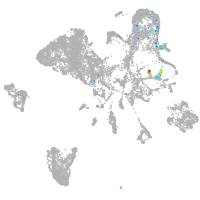
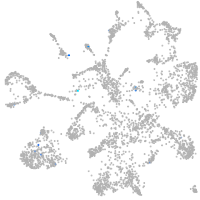
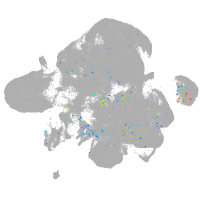
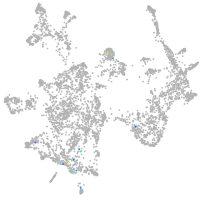
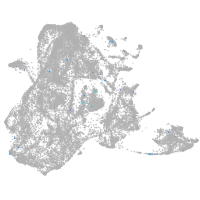
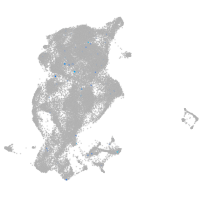
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.156 | hspb1 | -0.027 |
| rgs1 | 0.148 | cx43.4 | -0.025 |
| si:dkeyp-80c12.5 | 0.146 | ddx39ab | -0.025 |
| CABZ01072157.1 | 0.142 | hnrnpa1b | -0.025 |
| LO018550.2 | 0.142 | akap12b | -0.024 |
| FP016247.2 | 0.140 | hnrnpub | -0.024 |
| LOC103909697 | 0.136 | mki67 | -0.024 |
| LOC100536671 | 0.135 | seta | -0.024 |
| trpm5 | 0.133 | chaf1a | -0.023 |
| XLOC-039931 | 0.133 | dkc1 | -0.023 |
| BX005256.1 | 0.132 | nop58 | -0.023 |
| XLOC-001375 | 0.131 | ppm1g | -0.023 |
| avil | 0.129 | slbp | -0.023 |
| kcnk17 | 0.127 | sox11b | -0.023 |
| zgc:171929 | 0.126 | ssrp1a | -0.023 |
| gna14a | 0.125 | brd3a | -0.022 |
| BX005012.1 | 0.124 | hnrnph1l | -0.022 |
| zgc:158412 | 0.124 | lig1 | -0.022 |
| pkd1l3 | 0.121 | marcksb | -0.022 |
| si:ch73-199e17.1 | 0.120 | nono | -0.022 |
| gng13a | 0.118 | qkia | -0.022 |
| si:dkey-266f7.10 | 0.117 | snrpb | -0.022 |
| hepacam2 | 0.116 | taf15 | -0.022 |
| gyg1b | 0.114 | top1l | -0.022 |
| scinla | 0.112 | zgc:110216 | -0.022 |
| CR318588.3 | 0.111 | zgc:110425 | -0.022 |
| trpa1b | 0.110 | ccna2 | -0.021 |
| prr15la | 0.108 | fbxo5 | -0.021 |
| gbe1a | 0.107 | hdac1 | -0.021 |
| areg | 0.106 | ilf2 | -0.021 |
| calca | 0.106 | lbr | -0.021 |
| fgl2a | 0.106 | nasp | -0.021 |
| pitx1 | 0.106 | nucks1a | -0.021 |
| XLOC-041394 | 0.102 | ppig | -0.021 |
| CR759836.1 | 0.101 | rbbp4 | -0.021 |