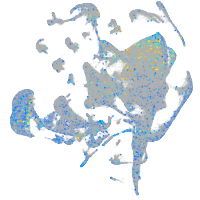
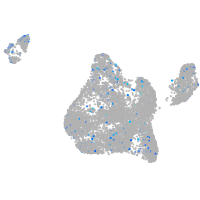
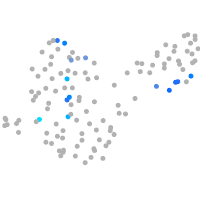
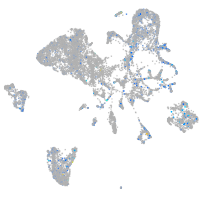
hypothetical protein FLJ11011-like (H. sapiens)
ZFIN 
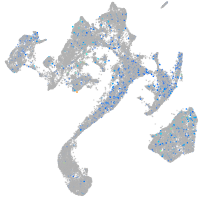
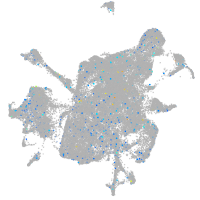
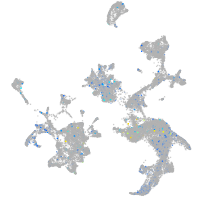
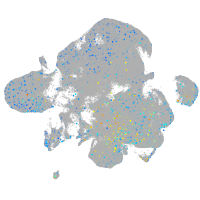
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpx9 | 0.071 | hmgb2b | -0.055 |
| mipa | 0.066 | si:ch211-222l21.1 | -0.053 |
| capn3a | 0.064 | hmgn2 | -0.050 |
| lim2.3 | 0.064 | h2afvb | -0.048 |
| cryba1l2 | 0.063 | hmga1a | -0.048 |
| lim2.4 | 0.063 | rrm1 | -0.045 |
| cryba1b | 0.061 | mdka | -0.045 |
| crybb1l2 | 0.060 | ahcy | -0.044 |
| cx23 | 0.060 | rplp0 | -0.044 |
| crybb1l1 | 0.060 | ran | -0.044 |
| endou2 | 0.059 | hmgb2a | -0.042 |
| crygmx | 0.058 | pcna | -0.042 |
| cryba1l1 | 0.058 | h3f3a | -0.041 |
| cryba4 | 0.058 | eef1b2 | -0.041 |
| cryba2b | 0.057 | ccna2 | -0.040 |
| cryba2a | 0.057 | stmn1a | -0.040 |
| mipb | 0.057 | rps23 | -0.040 |
| crybb1 | 0.057 | hnrnpa0l | -0.040 |
| lim2.1 | 0.056 | her15.1 | -0.039 |
| BFSP1 | 0.056 | rrm2 | -0.039 |
| crygn2 | 0.056 | dut | -0.039 |
| xirp1 | 0.055 | rack1 | -0.039 |
| si:ch211-254p10.2 | 0.055 | rplp1 | -0.038 |
| prx | 0.055 | rps9 | -0.038 |
| gapdhs | 0.055 | hes6 | -0.038 |
| aifm5 | 0.054 | rps7 | -0.038 |
| crygm2d8 | 0.053 | rpl38 | -0.038 |
| lctlb | 0.053 | lbr | -0.038 |
| sypb | 0.053 | rpl7a | -0.037 |
| plcd4a | 0.053 | mcm7 | -0.037 |
| crygm2d13 | 0.052 | hes2.2 | -0.037 |
| gja8b | 0.052 | rpl7 | -0.037 |
| atp6v0cb | 0.052 | mki67 | -0.037 |
| crybb1l3 | 0.052 | tspan7 | -0.037 |
| sncb | 0.051 | rplp2l | -0.037 |