"crystallin, beta B1, like 1"
ZFIN 
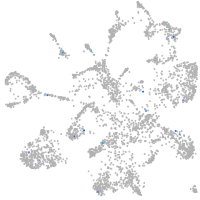
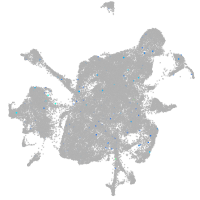
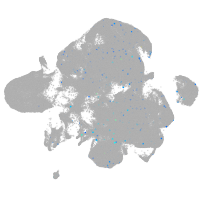
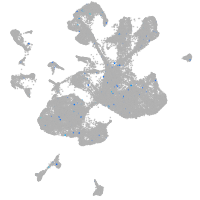
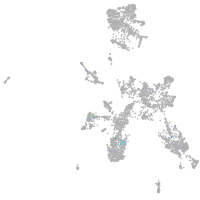
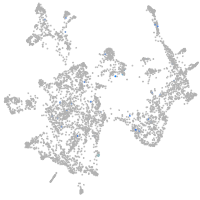
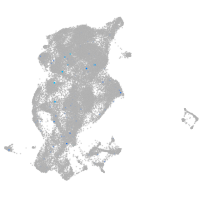
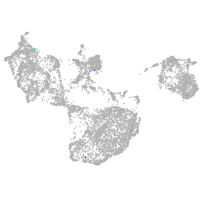
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mipb | 0.486 | sult1st6 | -0.025 |
| crygmx | 0.431 | mthfd1b | -0.024 |
| gja8b | 0.417 | galm | -0.024 |
| sall1b | 0.384 | dgat2 | -0.023 |
| lim2.4 | 0.380 | pla2g12b | -0.022 |
| mipa | 0.362 | ugt1a7 | -0.022 |
| cryba1b | 0.345 | ndufa8 | -0.022 |
| cx23 | 0.342 | gstr | -0.022 |
| gja8a | 0.337 | mccc1 | -0.022 |
| lim2.3 | 0.315 | creb3l3a | -0.021 |
| prox2 | 0.311 | cdo1 | -0.021 |
| crybb1l2 | 0.305 | zgc:163080 | -0.021 |
| crygm2e | 0.297 | b4galt1l | -0.021 |
| lim2.1 | 0.296 | si:ch211-212k18.7 | -0.021 |
| crygn2 | 0.292 | msrb2 | -0.021 |
| si:ch211-255g12.6 | 0.290 | zgc:136439 | -0.021 |
| grifin | 0.287 | tmem254 | -0.020 |
| cryba4 | 0.286 | faxdc2 | -0.020 |
| cryba1l1 | 0.274 | cyp3c1 | -0.020 |
| nrl | 0.237 | cideb | -0.020 |
| LOC110438002 | 0.203 | porb | -0.020 |
| nags | 0.201 | cyp2k16 | -0.020 |
| cryba2a | 0.198 | bco1 | -0.020 |
| crygm2d11 | 0.196 | agmo | -0.020 |
| s1pr3a | 0.196 | aldh8a1 | -0.020 |
| cryba1l2 | 0.186 | si:ch211-93g23.2 | -0.019 |
| crybb1 | 0.185 | abcg2a | -0.019 |
| XLOC-016003 | 0.179 | ethe1 | -0.019 |
| cryba2b | 0.178 | pck1 | -0.019 |
| BFSP1 | 0.173 | slc41a1 | -0.019 |
| BX545917.5 | 0.170 | ada | -0.019 |
| BX682558.1 | 0.162 | tkfc | -0.019 |
| pimr130 | 0.156 | PDPR | -0.019 |
| col28a1b | 0.151 | paip2b | -0.019 |
| otc | 0.145 | pon3.2 | -0.019 |