cell death inducing DFFA like effector b
ZFIN 
Other cell groups


all cells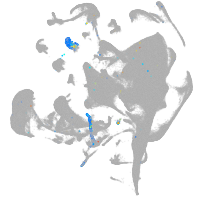 |
blastomeres |
PGCs |
endoderm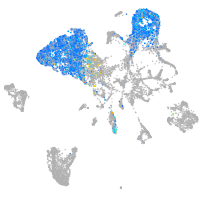 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 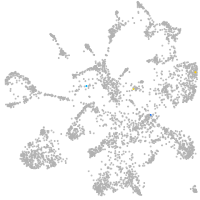 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 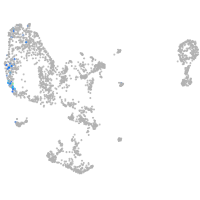 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pla2g12b | 0.663 | marcksl1a | -0.093 |
| apobb.1 | 0.659 | hmgb1b | -0.088 |
| rdh1 | 0.658 | hmgb3a | -0.086 |
| apoa4b.1 | 0.654 | tuba1c | -0.084 |
| cyp4v8 | 0.650 | jpt1b | -0.082 |
| cyp8b1 | 0.649 | nova2 | -0.079 |
| afp4 | 0.624 | hnrnpa0a | -0.078 |
| apoc2 | 0.614 | gpm6aa | -0.077 |
| slco1d1 | 0.613 | marcksb | -0.075 |
| fbp1b | 0.611 | atp6v0cb | -0.073 |
| sult3st2 | 0.608 | rtn1a | -0.073 |
| dgat2 | 0.606 | vdac1 | -0.073 |
| etnppl | 0.600 | gpm6ab | -0.071 |
| tdo2a | 0.599 | fam168a | -0.070 |
| creb3l3a | 0.598 | gapdhs | -0.070 |
| g6pca.2 | 0.594 | mdkb | -0.070 |
| mttp | 0.589 | tuba1a | -0.070 |
| tm4sf5 | 0.581 | rnasekb | -0.069 |
| cyp2p9 | 0.579 | tubb5 | -0.069 |
| gda | 0.574 | si:ch1073-429i10.3.1 | -0.069 |
| pklr | 0.573 | idh2 | -0.068 |
| rltgr | 0.571 | tmeff1b | -0.068 |
| ugt5b4 | 0.571 | celf2 | -0.067 |
| pnp4b | 0.569 | atrx | -0.066 |
| cx32.3 | 0.568 | CABZ01080702.1 | -0.066 |
| golt1a | 0.560 | fxyd6l | -0.066 |
| tmem86b | 0.560 | nucks1a | -0.066 |
| slc27a2a | 0.559 | si:ch211-133n4.4 | -0.066 |
| f7 | 0.557 | gnb1b | -0.065 |
| serpinf2a | 0.556 | h2afva | -0.065 |
| si:ch211-288g17.4 | 0.553 | oaz1b | -0.065 |
| ugt5b3 | 0.553 | chd4a | -0.064 |
| a2ml | 0.552 | si:ch211-288g17.3 | -0.064 |
| si:ch211-284e20.8 | 0.552 | si:ch73-46j18.5 | -0.064 |
| shbg | 0.548 | elavl3 | -0.063 |




