bone morphogenetic protein 2a
ZFIN 
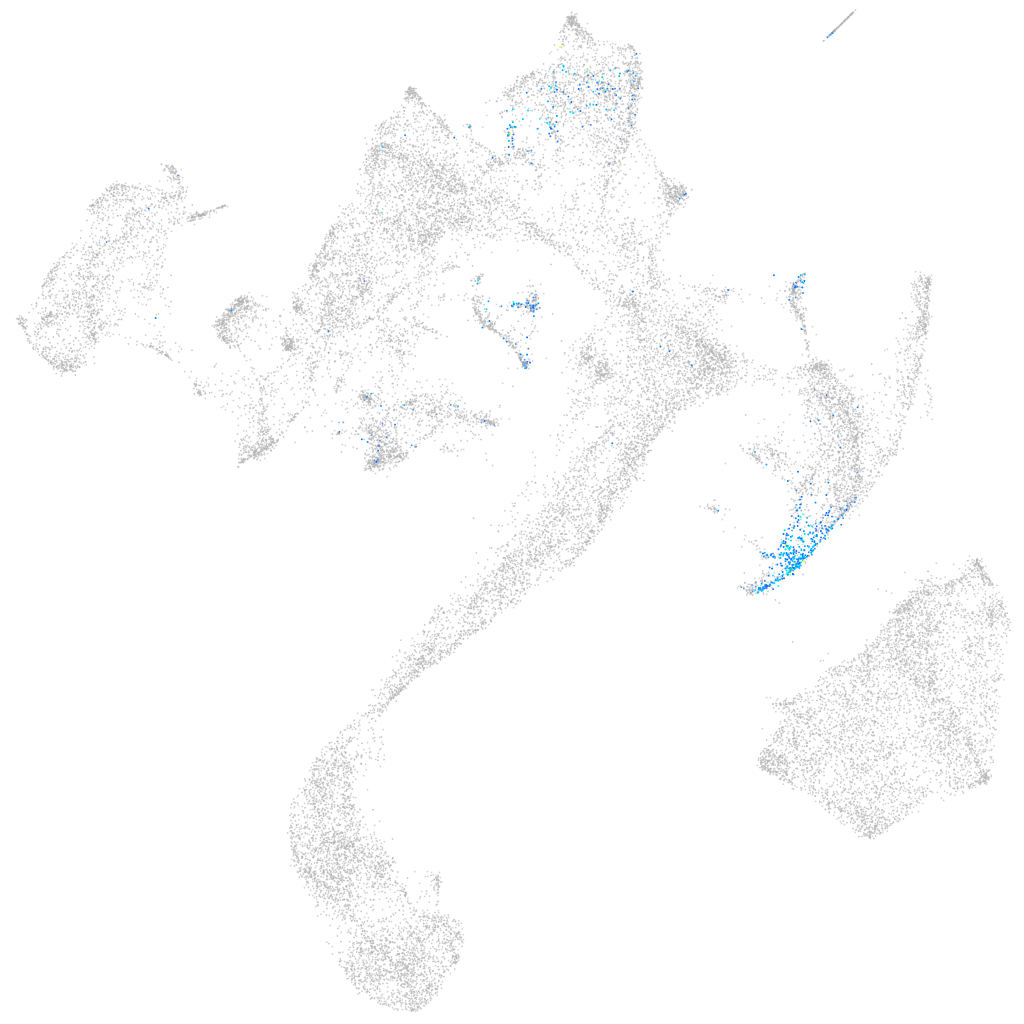
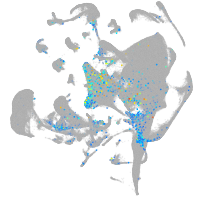
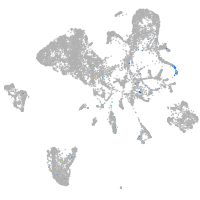
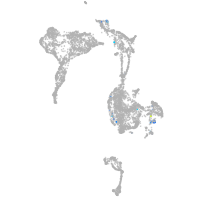
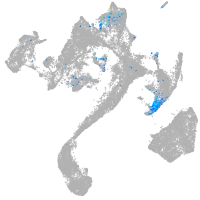
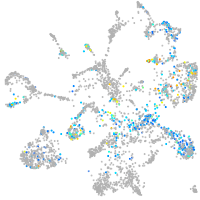
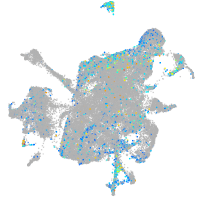
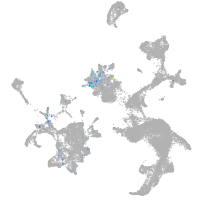
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fgf3 | 0.369 | ttn.1 | -0.107 |
| gdf11 | 0.320 | hsp90aa1.1 | -0.101 |
| hoxd12a | 0.292 | actc1b | -0.099 |
| tagln3b | 0.283 | ttn.2 | -0.098 |
| hoxa13b | 0.280 | fxr2 | -0.089 |
| tll1 | 0.276 | pabpc4 | -0.088 |
| thbs2a | 0.273 | tmem38a | -0.086 |
| hoxd10a | 0.271 | aldoab | -0.085 |
| hoxb10a | 0.265 | zgc:92429 | -0.085 |
| cyp26a1 | 0.249 | klhl41b | -0.084 |
| enc3 | 0.242 | fabp3 | -0.084 |
| sall4 | 0.234 | atp2a1 | -0.084 |
| fgf8a | 0.232 | ckma | -0.083 |
| tbxtb | 0.221 | ak1 | -0.083 |
| esrrga | 0.220 | ckmb | -0.083 |
| XLOC-003690 | 0.220 | gatm | -0.082 |
| itm2cb | 0.219 | mybphb | -0.081 |
| her15.1 | 0.210 | desma | -0.080 |
| hoxa10b | 0.209 | acta1a | -0.080 |
| depdc7a | 0.207 | gapdh | -0.080 |
| ephb4b | 0.207 | tpma | -0.080 |
| b3gnt7l | 0.206 | srl | -0.079 |
| zgc:162939 | 0.205 | tnnc2 | -0.079 |
| greb1 | 0.204 | zgc:92518 | -0.078 |
| NAV1 (1 of many) | 0.200 | CABZ01078594.1 | -0.078 |
| her12 | 0.199 | neb | -0.078 |
| hoxd9a | 0.190 | cav3 | -0.078 |
| tbxta | 0.190 | tnni2b.1 | -0.077 |
| adam8a | 0.189 | actc1a | -0.076 |
| bambia | 0.187 | txlnbb | -0.076 |
| CABZ01076996.1 | 0.182 | gamt | -0.076 |
| wnt2bb | 0.182 | cox17 | -0.075 |
| hoxd3a | 0.182 | ldb3a | -0.075 |
| hoxc11a | 0.181 | zgc:101853 | -0.074 |
| hoxa11a | 0.179 | cox7c | -0.074 |