"UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like"
ZFIN 
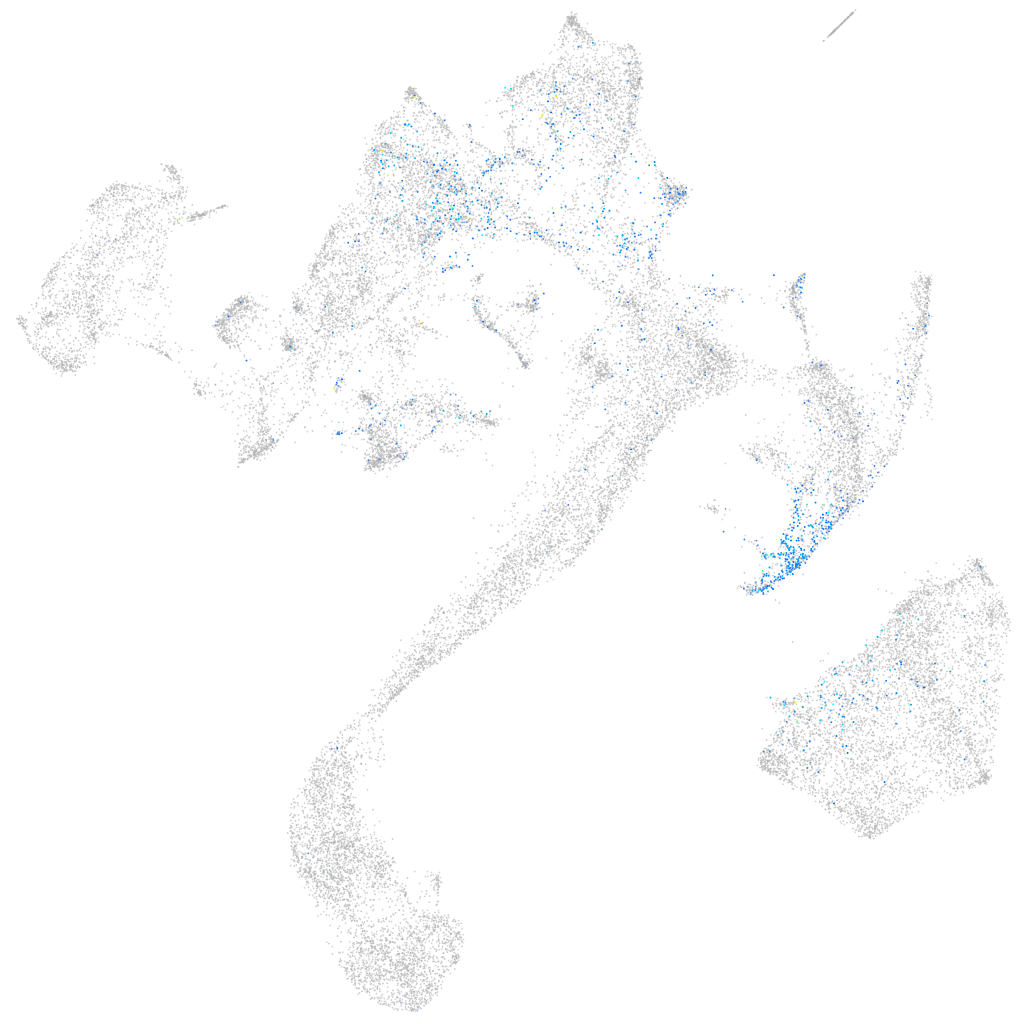
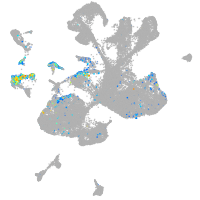
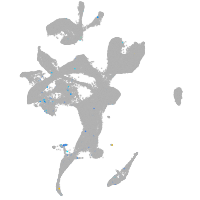
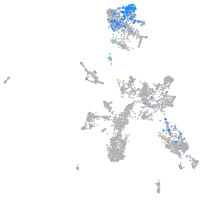
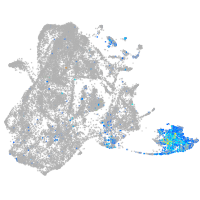
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fgf3 | 0.235 | ttn.1 | -0.128 |
| gdf11 | 0.220 | ttn.2 | -0.117 |
| bmp2a | 0.206 | hsp90aa1.1 | -0.112 |
| thbs2a | 0.199 | actc1b | -0.109 |
| hoxd12a | 0.199 | aldoab | -0.105 |
| tagln3b | 0.195 | pabpc4 | -0.105 |
| cyp26a1 | 0.193 | ak1 | -0.104 |
| tll1 | 0.185 | tmem38a | -0.103 |
| hoxa13b | 0.169 | atp2a1 | -0.102 |
| zgc:162939 | 0.169 | ckmb | -0.102 |
| hoxd10a | 0.163 | gapdh | -0.102 |
| fgf8a | 0.162 | klhl41b | -0.101 |
| wls | 0.158 | fxr2 | -0.101 |
| itm2cb | 0.155 | srl | -0.101 |
| hoxb10a | 0.152 | ckma | -0.100 |
| ephb4b | 0.151 | mybphb | -0.100 |
| enc3 | 0.151 | tnnc2 | -0.099 |
| id3 | 0.146 | desma | -0.099 |
| sall4 | 0.146 | neb | -0.097 |
| rarga | 0.143 | CABZ01078594.1 | -0.096 |
| hoxa10b | 0.140 | ldb3a | -0.096 |
| hoxc11a | 0.139 | cox17 | -0.096 |
| ptmaa | 0.136 | cav3 | -0.096 |
| XLOC-003690 | 0.136 | acta1b | -0.095 |
| tbxtb | 0.135 | tpma | -0.095 |
| depdc7a | 0.134 | acta1a | -0.095 |
| CABZ01076996.1 | 0.133 | zgc:101853 | -0.094 |
| dand5 | 0.133 | actc1a | -0.093 |
| nradd | 0.132 | ldb3b | -0.092 |
| adam8a | 0.131 | txlnbb | -0.092 |
| her15.1 | 0.130 | gatm | -0.092 |
| tubb4b | 0.130 | atp5if1b | -0.091 |
| h3f3a | 0.129 | actn3b | -0.091 |
| esrrga | 0.129 | CABZ01072309.1 | -0.091 |
| her12 | 0.129 | eno1a | -0.091 |