zona pellucida-like domain containing 1b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm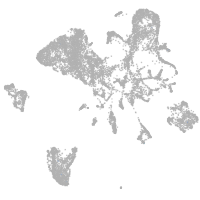 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
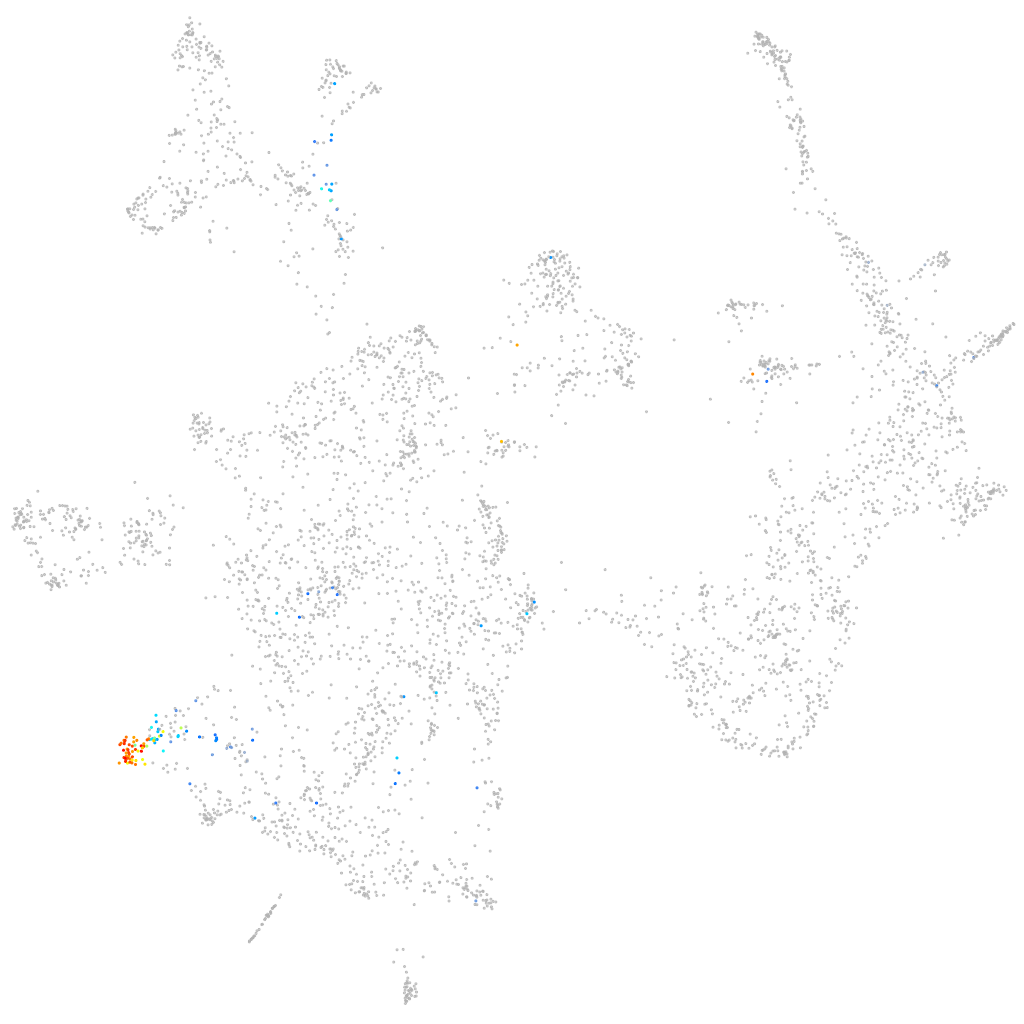
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zpld1a | 0.752 | krt8 | -0.103 |
| hhla1 | 0.617 | tubb2b | -0.099 |
| msx3 | 0.581 | tuba8l4 | -0.090 |
| FAM163A | 0.543 | marcksl1b | -0.085 |
| adcy2b | 0.446 | krt18a.1 | -0.085 |
| cyp26b1 | 0.436 | fkbp1aa | -0.082 |
| slc13a5a | 0.435 | hmgb2a | -0.079 |
| otog | 0.415 | cldnh | -0.078 |
| mia | 0.382 | stmn1a | -0.077 |
| nfasca | 0.378 | snrpe | -0.077 |
| dlx1a | 0.374 | zgc:158343 | -0.076 |
| nog2 | 0.372 | gsnb | -0.075 |
| znf536 | 0.368 | npm1a | -0.075 |
| adam8a | 0.368 | snrpf | -0.074 |
| dlx6a | 0.362 | snrpd1 | -0.074 |
| ncam2 | 0.355 | tma7 | -0.074 |
| cxcl14 | 0.353 | tmsb | -0.073 |
| kcnq5a | 0.352 | six2b | -0.073 |
| glula | 0.347 | pcna | -0.073 |
| slc25a22b | 0.344 | dlx3b | -0.073 |
| sulf2a | 0.344 | pax2a | -0.072 |
| cfd | 0.340 | krt15 | -0.072 |
| isl1 | 0.332 | si:ch211-195b11.3 | -0.071 |
| ahcyl1 | 0.330 | nqo1 | -0.071 |
| trim101 | 0.326 | tfap2a | -0.070 |
| dlx2a | 0.323 | hnrnpaba | -0.070 |
| myo18aa | 0.322 | pvalb8 | -0.069 |
| chl1b | 0.315 | nutf2l | -0.068 |
| smad9 | 0.310 | cyt1 | -0.068 |
| cryabb | 0.309 | foxg1a | -0.068 |
| dlx5a | 0.306 | dkc1 | -0.067 |
| BX663503.3 | 0.301 | gpc1a | -0.067 |
| slc16a1a | 0.301 | ptges3b | -0.067 |
| ifit10 | 0.301 | krt4 | -0.067 |
| adgrg3 | 0.300 | cx43.4 | -0.067 |