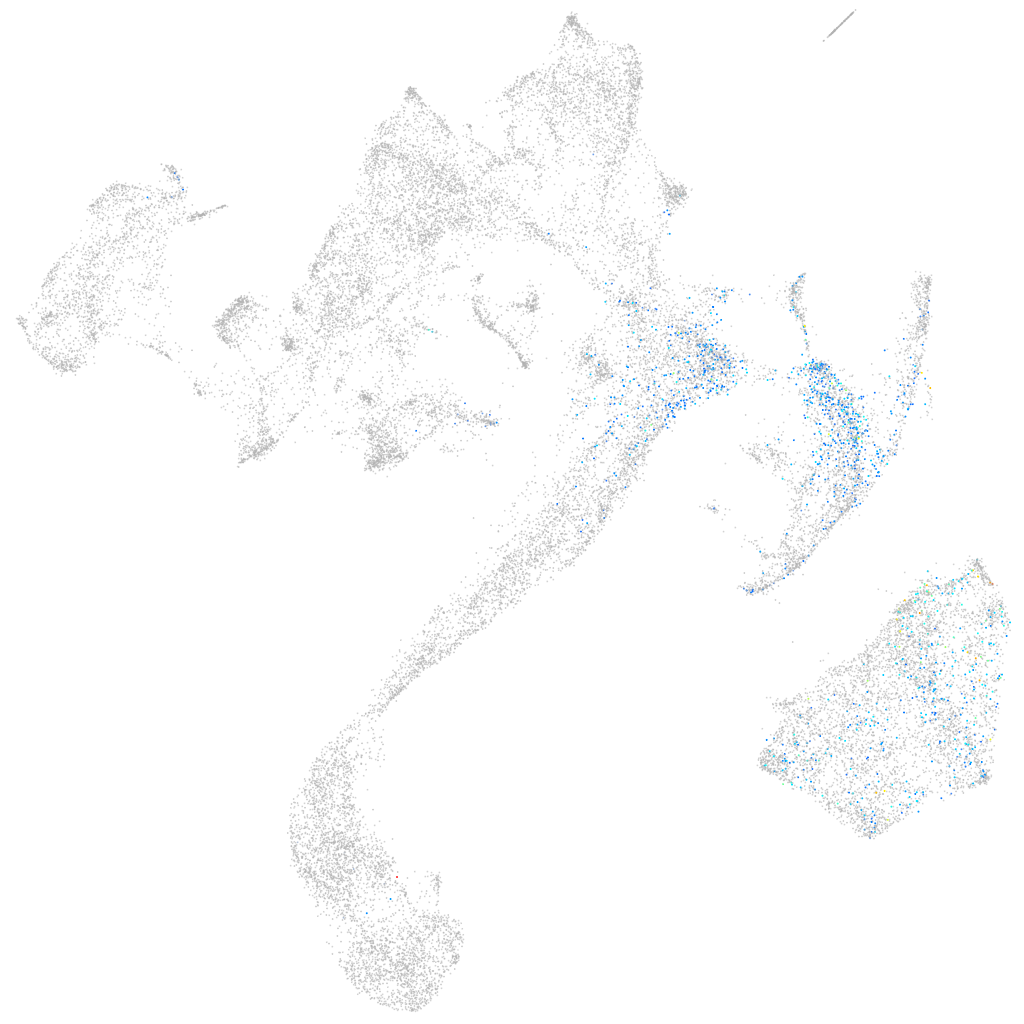
zinc finger-like gene 1h
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoc1 | 0.201 | fabp3 | -0.152 |
| tbx6 | 0.192 | actc1b | -0.135 |
| myf5 | 0.183 | bhmt | -0.125 |
| efnb2b | 0.179 | eef1da | -0.122 |
| apoeb | 0.177 | sparc | -0.115 |
| fn1b | 0.175 | ak1 | -0.115 |
| cx43.4 | 0.174 | ckma | -0.115 |
| nid2a | 0.173 | ckmb | -0.114 |
| pcdh8 | 0.168 | atp2a1 | -0.114 |
| draxin | 0.164 | pabpc4 | -0.113 |
| gnaia | 0.163 | eno1a | -0.113 |
| nop58 | 0.163 | aldoab | -0.112 |
| npm1a | 0.160 | tmem38a | -0.111 |
| grin2da | 0.158 | mylpfa | -0.110 |
| ncl | 0.157 | eif4a1b | -0.109 |
| si:ch73-281n10.2 | 0.157 | tpi1b | -0.108 |
| syncrip | 0.155 | pmp22a | -0.107 |
| BX001014.2 | 0.155 | gamt | -0.106 |
| BX927258.1 | 0.155 | col1a2 | -0.106 |
| nop56 | 0.154 | acta1b | -0.105 |
| hes6 | 0.151 | tnnc2 | -0.105 |
| znfl1k | 0.150 | srl | -0.103 |
| fbl | 0.149 | rpl37 | -0.103 |
| asph | 0.149 | ndrg2 | -0.102 |
| XLOC-004280 | 0.148 | vdac3 | -0.100 |
| crabp2b | 0.148 | gapdh | -0.100 |
| dlc | 0.148 | myl1 | -0.100 |
| hmga1a | 0.148 | col1a1a | -0.100 |
| ilf3b | 0.148 | ttn.2 | -0.100 |
| her11 | 0.148 | neb | -0.099 |
| dkc1 | 0.148 | pvalb1 | -0.099 |
| nucks1a | 0.147 | desma | -0.099 |
| ebna1bp2 | 0.147 | eno3 | -0.099 |
| BX005254.3 | 0.146 | tpma | -0.099 |
| msgn1 | 0.146 | pvalb2 | -0.098 |