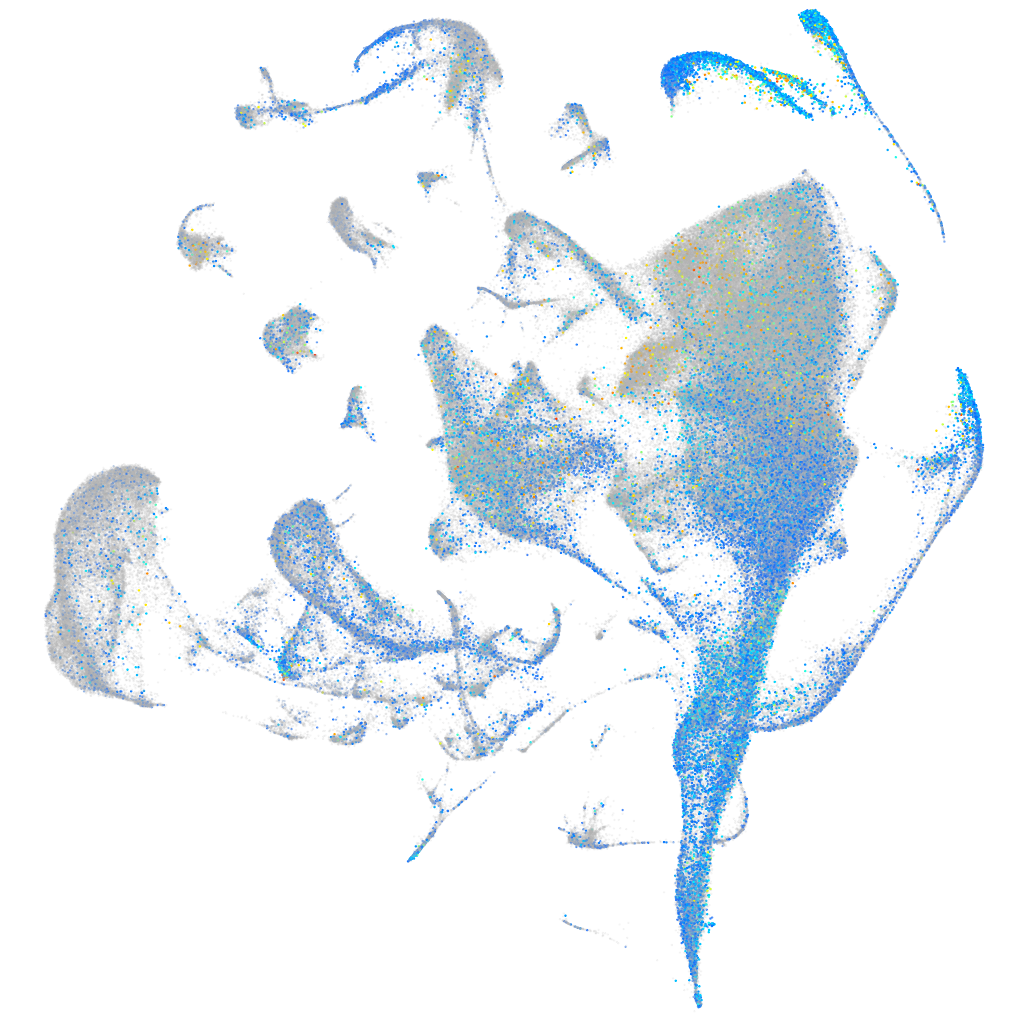zinc finger protein 106a
ZFIN 
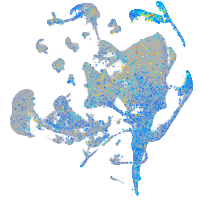
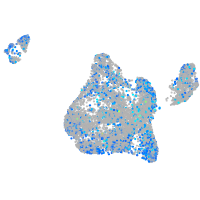
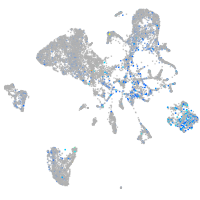
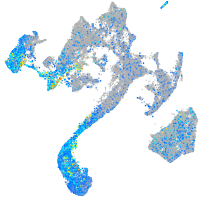
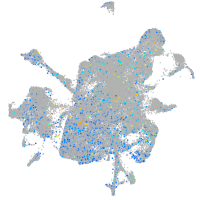
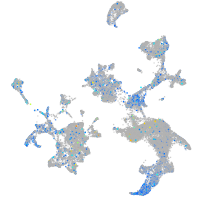
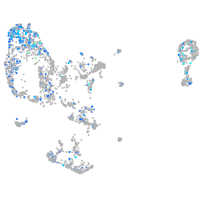
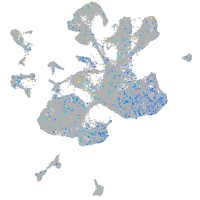
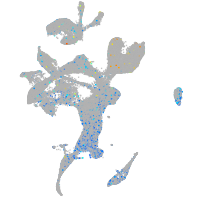
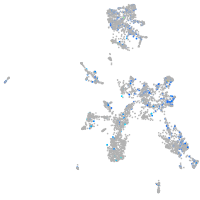
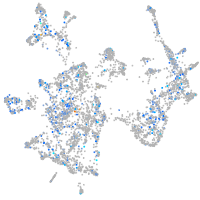
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| srl | 0.254 | ckbb | -0.090 |
| casq2 | 0.253 | gpm6aa | -0.088 |
| cav3 | 0.249 | rnasekb | -0.088 |
| desma | 0.245 | gpm6ab | -0.085 |
| cavin4a | 0.242 | atp6v0cb | -0.084 |
| cavin4b | 0.239 | gapdhs | -0.080 |
| CABZ01072309.1 | 0.236 | stmn1b | -0.080 |
| aldoab | 0.235 | tuba1c | -0.080 |
| ldb3b | 0.235 | elavl3 | -0.075 |
| si:ch211-266g18.10 | 0.234 | sncb | -0.075 |
| trdn | 0.234 | calm1a | -0.074 |
| actn3b | 0.232 | ccni | -0.073 |
| neb | 0.230 | vamp2 | -0.073 |
| klhl43 | 0.230 | atp6v1e1b | -0.072 |
| pgam2 | 0.230 | krt4 | -0.071 |
| tmem182a | 0.229 | atpv0e2 | -0.069 |
| klhl31 | 0.228 | gng3 | -0.069 |
| zgc:158296 | 0.228 | rtn1b | -0.067 |
| casq1b | 0.226 | atp6v1g1 | -0.066 |
| myom1a | 0.225 | cyt1 | -0.065 |
| tmem38a | 0.223 | elavl4 | -0.065 |
| stac3 | 0.222 | atp6ap2 | -0.064 |
| CABZ01078594.1 | 0.221 | si:dkeyp-75h12.5 | -0.064 |
| pabpc4 | 0.220 | marcksl1a | -0.063 |
| CU681836.1 | 0.219 | si:ch211-195b11.3 | -0.062 |
| ldb3a | 0.219 | cspg5a | -0.061 |
| prr33 | 0.218 | icn | -0.061 |
| atp1a2a | 0.217 | stx1b | -0.061 |
| CR376766.2 | 0.217 | cyt1l | -0.060 |
| dhrs7cb | 0.216 | icn2 | -0.060 |
| myot | 0.216 | ip6k2a | -0.060 |
| prx | 0.215 | nsg2 | -0.060 |
| jph2 | 0.215 | stxbp1a | -0.060 |
| hhatla | 0.214 | atp1a3a | -0.059 |
| txlnbb | 0.214 | olfm1b | -0.059 |