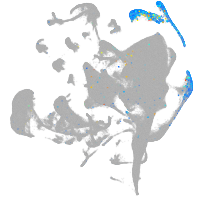
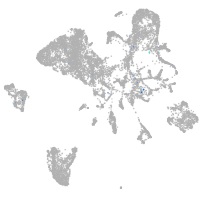
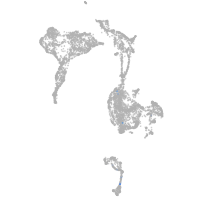
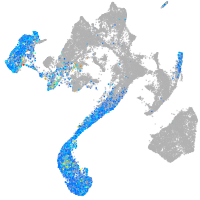
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| srl | 0.612 | hmgb1b | -0.098 |
| aldoab | 0.574 | ywhaqb | -0.097 |
| klhl31 | 0.572 | marcksl1a | -0.097 |
| cav3 | 0.570 | si:ch211-288g17.3 | -0.093 |
| desma | 0.570 | tuba1c | -0.088 |
| txlnbb | 0.566 | gapdhs | -0.085 |
| tmem182a | 0.564 | gnb1b | -0.085 |
| CABZ01078594.1 | 0.563 | gpm6aa | -0.085 |
| tmem38a | 0.560 | ckbb | -0.084 |
| CABZ01072309.1 | 0.555 | nova2 | -0.084 |
| ldb3b | 0.551 | h2afva | -0.083 |
| casq2 | 0.550 | rnasekb | -0.083 |
| actn3b | 0.548 | chd4a | -0.082 |
| ldb3a | 0.545 | cbx5 | -0.079 |
| zgc:158296 | 0.542 | gpm6ab | -0.078 |
| mybphb | 0.539 | mdkb | -0.078 |
| trdn | 0.539 | pfn2 | -0.078 |
| cavin4b | 0.537 | anp32a | -0.076 |
| neb | 0.535 | ddx5 | -0.076 |
| hhatla | 0.530 | atrx | -0.075 |
| klhl43 | 0.528 | calm3b | -0.075 |
| pgam2 | 0.528 | ctsla | -0.075 |
| acta1a | 0.527 | zc4h2 | -0.074 |
| stac3 | 0.525 | atp6v0cb | -0.073 |
| txlnba | 0.524 | seta | -0.073 |
| cacng1a | 0.523 | tmem258 | -0.073 |
| cavin4a | 0.517 | tuba1a | -0.073 |
| myom1a | 0.517 | ywhabb | -0.073 |
| actn3a | 0.514 | ywhaqa | -0.073 |
| si:ch211-266g18.10 | 0.510 | hdac1 | -0.071 |
| smyd1a | 0.510 | hmgn3 | -0.070 |
| cacna1sb | 0.508 | marcksb | -0.070 |
| prr33 | 0.508 | pdia3 | -0.070 |
| actc1a | 0.506 | tubb5 | -0.070 |
| atp1a2a | 0.504 | calm1a | -0.070 |