kelch-like family member 31
ZFIN 
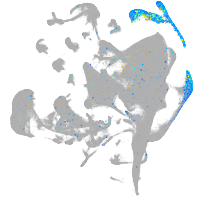
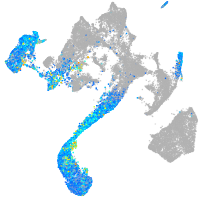
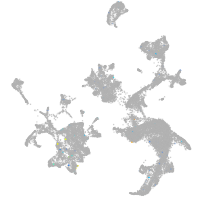
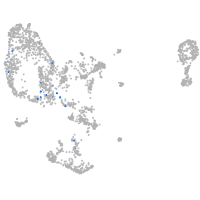
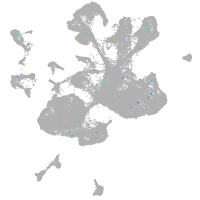
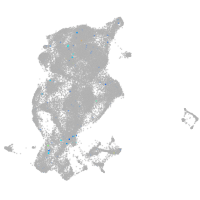
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| srl | 0.653 | hmgb1b | -0.106 |
| desma | 0.642 | marcksl1a | -0.104 |
| txlnbb | 0.642 | si:ch211-288g17.3 | -0.100 |
| cav3 | 0.610 | ywhaqb | -0.099 |
| txlnba | 0.608 | chd4a | -0.090 |
| aldoab | 0.607 | tuba1c | -0.089 |
| tmem38a | 0.594 | cbx5 | -0.087 |
| acta1a | 0.590 | gapdhs | -0.086 |
| CABZ01078594.1 | 0.585 | gnb1b | -0.086 |
| CABZ01072309.1 | 0.578 | gpm6aa | -0.085 |
| actc1a | 0.576 | h2afva | -0.085 |
| cacng1a | 0.572 | nova2 | -0.085 |
| CR376766.2 | 0.572 | ckbb | -0.083 |
| tmem182a | 0.572 | pfn2 | -0.082 |
| tnni2b.1 | 0.570 | anp32a | -0.081 |
| ldb3a | 0.569 | ctsla | -0.081 |
| mybphb | 0.567 | mdkb | -0.081 |
| casq2 | 0.566 | seta | -0.081 |
| ldb3b | 0.557 | atrx | -0.080 |
| rbfox1l | 0.553 | rnasekb | -0.080 |
| zgc:101853 | 0.550 | ddx5 | -0.079 |
| trdn | 0.550 | gpm6ab | -0.078 |
| zgc:158296 | 0.550 | tuba1a | -0.078 |
| actn3b | 0.549 | ywhaqa | -0.078 |
| smyd1b | 0.547 | tmem258 | -0.077 |
| stac3 | 0.545 | zc4h2 | -0.077 |
| neb | 0.544 | hmgn3 | -0.075 |
| cavin4b | 0.541 | pdia3 | -0.075 |
| chrnd | 0.541 | si:ch211-137a8.4 | -0.075 |
| apobec2a | 0.537 | calm3b | -0.074 |
| zgc:92518 | 0.536 | morf4l1 | -0.073 |
| klhl43 | 0.534 | stmn1a | -0.073 |
| fitm1 | 0.533 | tubb5 | -0.073 |
| tnnt2c | 0.533 | ywhabb | -0.073 |
| klhl41b | 0.528 | hdac1 | -0.072 |