zinc finger protein 106a
ZFIN 
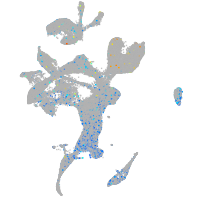
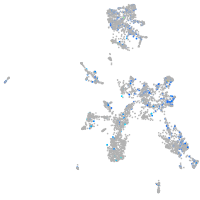
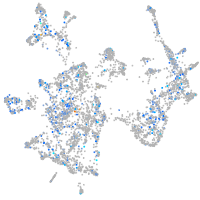
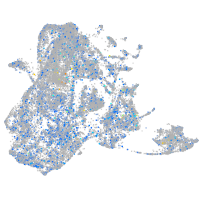
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp2a1 | 0.562 | hsp90ab1 | -0.509 |
| neb | 0.548 | h3f3d | -0.463 |
| gapdh | 0.546 | ran | -0.434 |
| casq2 | 0.546 | h2afvb | -0.419 |
| prx | 0.540 | pabpc1a | -0.418 |
| si:ch211-266g18.10 | 0.539 | actb2 | -0.415 |
| ckmb | 0.538 | cirbpb | -0.412 |
| ckma | 0.535 | si:ch73-1a9.3 | -0.410 |
| aldoab | 0.522 | ubc | -0.407 |
| ldb3b | 0.518 | hmgb2b | -0.404 |
| ak1 | 0.518 | naca | -0.391 |
| tnnt3a | 0.515 | hnrnpaba | -0.390 |
| myom1a | 0.514 | fthl27 | -0.389 |
| cav3 | 0.511 | hmgn2 | -0.388 |
| ttn.2 | 0.509 | khdrbs1a | -0.388 |
| actn3b | 0.507 | si:ch211-222l21.1 | -0.376 |
| CABZ01072309.1 | 0.507 | ptmab | -0.366 |
| ank1a | 0.504 | si:ch73-281n10.2 | -0.364 |
| srl | 0.503 | hmgb2a | -0.362 |
| cavin4b | 0.501 | hmgn7 | -0.356 |
| cavin4a | 0.500 | hmga1a | -0.355 |
| pgam2 | 0.497 | cfl1 | -0.354 |
| trdn | 0.497 | hnrnpabb | -0.352 |
| tnnc2 | 0.496 | cirbpa | -0.350 |
| si:ch73-367p23.2 | 0.495 | eef1a1l1 | -0.349 |
| pabpc4 | 0.495 | hnrnpa0b | -0.343 |
| tmem38a | 0.493 | tuba8l4 | -0.342 |
| XLOC-001975 | 0.491 | sumo3a | -0.339 |
| ttn.1 | 0.489 | hmgn6 | -0.339 |
| eno3 | 0.489 | actb1 | -0.339 |
| XLOC-025819 | 0.488 | h2afva | -0.338 |
| nme2b.2 | 0.486 | snrpf | -0.336 |
| desma | 0.486 | cbx3a | -0.336 |
| klhl43 | 0.482 | setb | -0.329 |
| actc1b | 0.480 | sub1a | -0.325 |