zgc:92907
ZFIN 
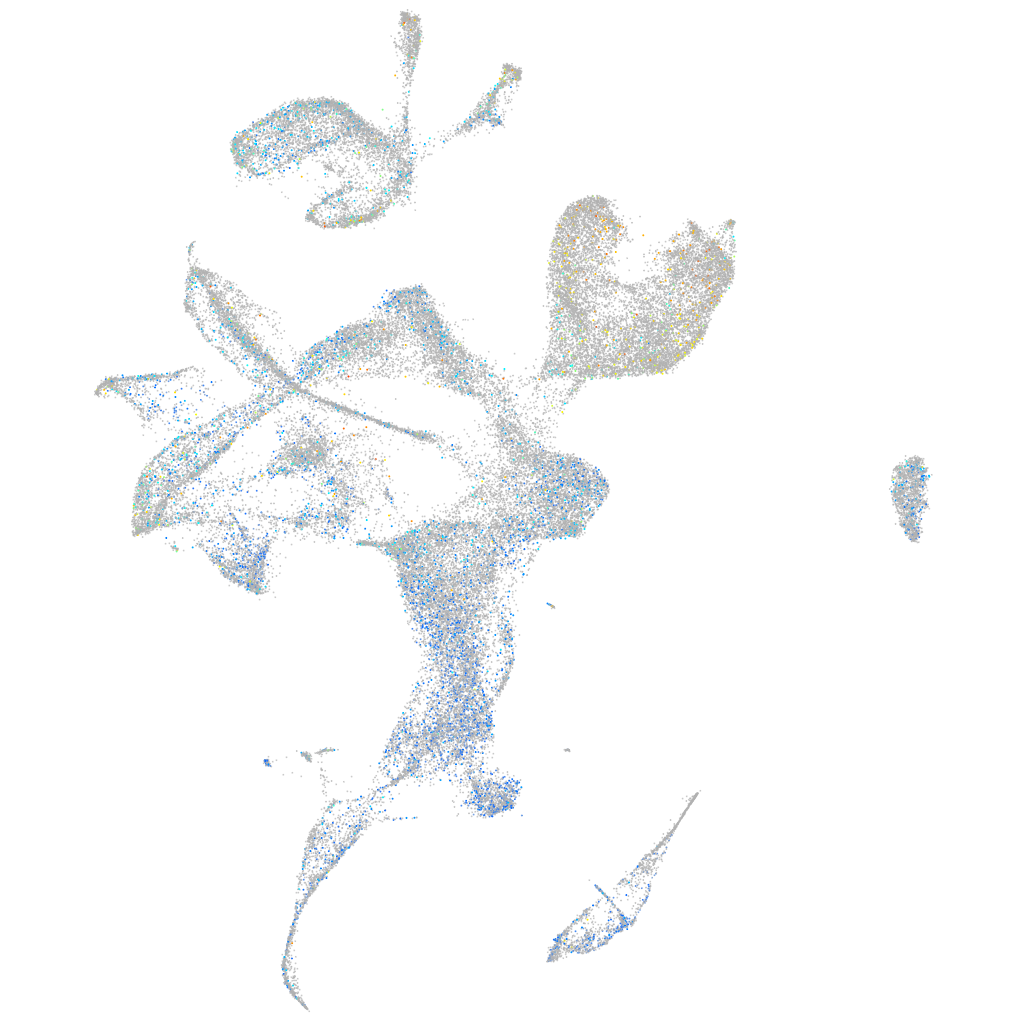
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5mc1 | 0.050 | hunk | -0.018 |
| atp5mc3b | 0.048 | scrt2 | -0.018 |
| uqcrc1 | 0.048 | crybb1l2 | -0.018 |
| gpx4b | 0.047 | prdm1b | -0.018 |
| ndufb9 | 0.047 | btg1 | -0.018 |
| atp5fa1 | 0.046 | crygm2d13 | -0.018 |
| ldhba | 0.046 | crybb1l1 | -0.018 |
| atp5f1b | 0.046 | si:ch211-222l21.1 | -0.017 |
| atp5f1c | 0.045 | cryba1b | -0.017 |
| ndufb3 | 0.044 | lim2.4 | -0.017 |
| cox8a | 0.044 | crygn2 | -0.017 |
| cox4i1 | 0.044 | crygmx | -0.017 |
| COX5B | 0.043 | cryba1l2 | -0.017 |
| mpc2 | 0.043 | mipb | -0.016 |
| atp5pb | 0.043 | grifin | -0.016 |
| ndufb8 | 0.042 | cryba2b | -0.016 |
| sdhb | 0.042 | crygm2d10 | -0.016 |
| ndufc2 | 0.040 | capn3a | -0.016 |
| ndufs4 | 0.040 | gadd45ga | -0.016 |
| ndufa10 | 0.040 | cryba2a | -0.016 |
| cox7a2a | 0.040 | mipa | -0.016 |
| ap2m1a | 0.040 | lim2.3 | -0.016 |
| cox5ab | 0.040 | crygm2d8 | -0.016 |
| cox6a1 | 0.039 | cryaa | -0.016 |
| si:ch1073-325m22.2 | 0.039 | si:ch211-255g12.6 | -0.016 |
| atp5pf | 0.039 | crygm2d12 | -0.015 |
| ndufa8 | 0.039 | lim2.5 | -0.015 |
| atp5f1d | 0.039 | crygm2d2 | -0.015 |
| ndufv2 | 0.039 | mylpfa | -0.015 |
| mpc1 | 0.039 | zswim5 | -0.015 |
| mif | 0.038 | gpx9 | -0.015 |
| arl3l1 | 0.038 | cryba4 | -0.014 |
| uqcrfs1 | 0.038 | zbtb20 | -0.014 |
| atp6v1g1 | 0.038 | crygmxl2 | -0.014 |
| suclg1 | 0.038 | crygm2d3 | -0.014 |