zgc:194551
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 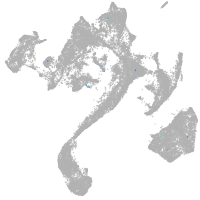 |
mural cells / non-skeletal muscle  |
mesenchyme 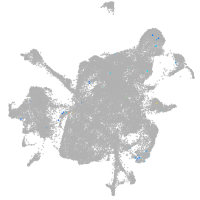 |
hematopoietic / vasculature 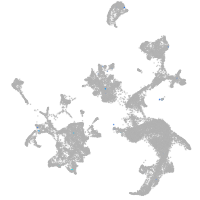 |
pronephros 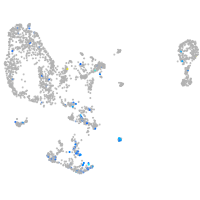 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin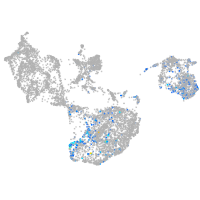 |
ionocytes / mucous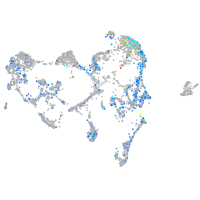 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sp6 | 0.284 | bhmt | -0.131 |
| krt97 | 0.244 | c1qtnf5 | -0.125 |
| zgc:174938 | 0.243 | fbln1 | -0.113 |
| alpi.1 | 0.237 | hgd | -0.113 |
| frem2a | 0.226 | pcbd1 | -0.111 |
| si:ch211-243g18.2 | 0.226 | cd82a | -0.108 |
| col14a1a | 0.222 | twist1a | -0.108 |
| vcana | 0.221 | rplp1 | -0.106 |
| slc7a11 | 0.206 | marcksl1a | -0.105 |
| sp9 | 0.205 | ttc36 | -0.105 |
| f11r.1 | 0.203 | tmem119b | -0.104 |
| cxcr4a | 0.201 | si:dkey-151g10.6 | -0.102 |
| tcf7 | 0.197 | prrx1a | -0.100 |
| phlda2 | 0.193 | prrx1b | -0.100 |
| emx3 | 0.193 | hpdb | -0.096 |
| cd9b | 0.192 | qdpra | -0.095 |
| cxcr4b | 0.189 | ptx3a | -0.095 |
| egfl6 | 0.189 | dcn | -0.095 |
| icn | 0.185 | nme2b.1 | -0.095 |
| bhlha9 | 0.184 | gpc1a | -0.092 |
| lama5 | 0.182 | cfl1 | -0.092 |
| cnpy1 | 0.182 | rpl37 | -0.092 |
| krt8 | 0.179 | col5a1 | -0.091 |
| net1 | 0.178 | fah | -0.089 |
| vamp8 | 0.178 | reck | -0.089 |
| fras1 | 0.178 | cd81a | -0.088 |
| apoc1 | 0.178 | slc16a10 | -0.087 |
| plekhh1 | 0.178 | twist1b | -0.086 |
| anxa5b | 0.177 | crabp2a | -0.086 |
| s100z | 0.177 | fmoda | -0.084 |
| epcam | 0.177 | twist3 | -0.084 |
| cldni | 0.176 | tmem176 | -0.084 |
| angptl7 | 0.174 | mab21l2 | -0.083 |
| tagln2 | 0.173 | pah | -0.082 |
| s100a10b | 0.172 | tgfbi | -0.082 |




