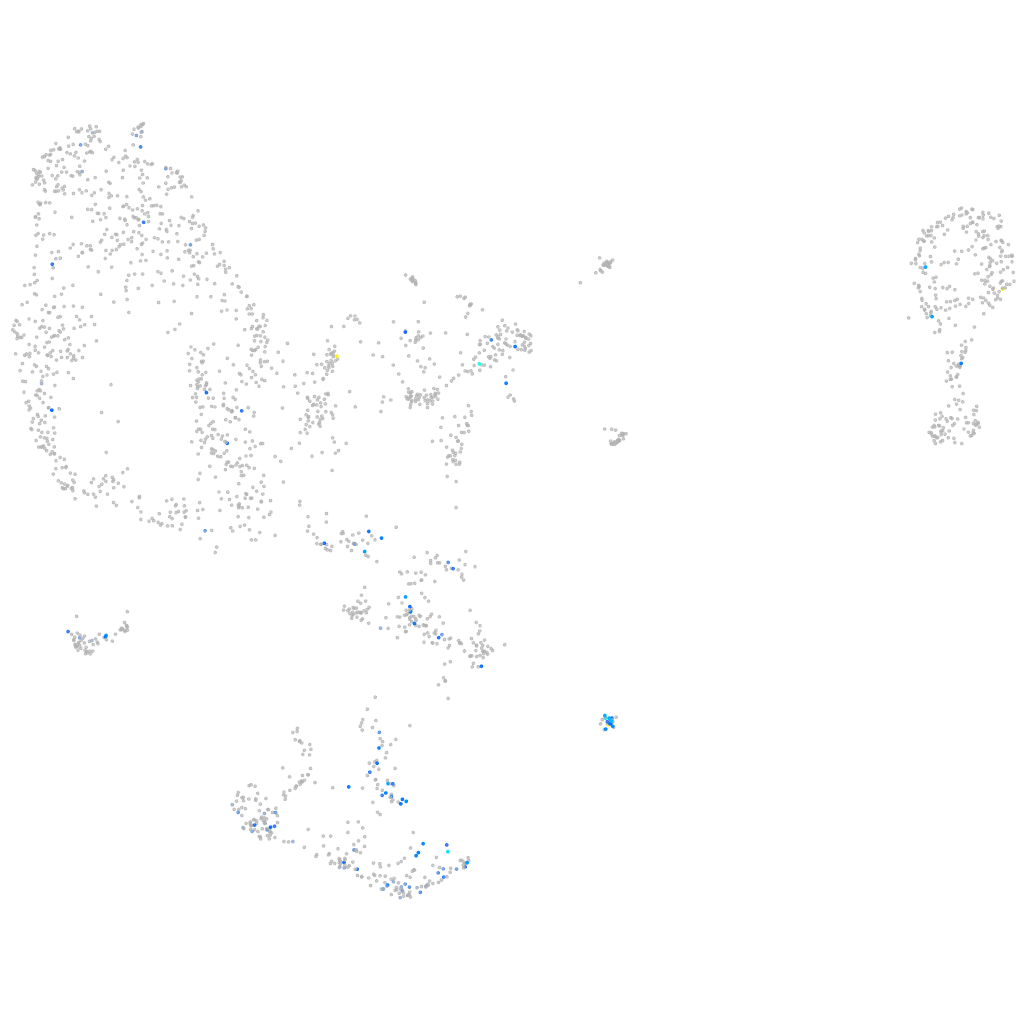
zgc:194551
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 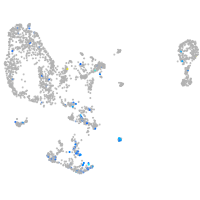 |
neural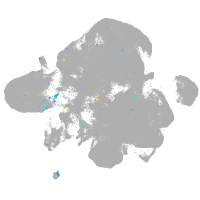 |
spinal cord / glia |
eye |
taste / olfactory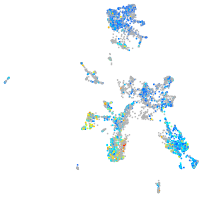 |
otic / lateral line |
epidermis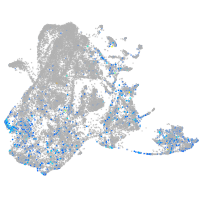 |
periderm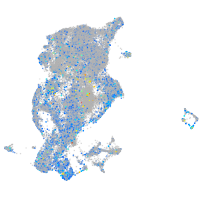 |
fin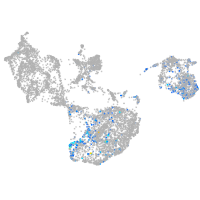 |
ionocytes / mucous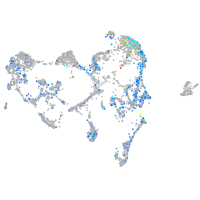 |
pigment cells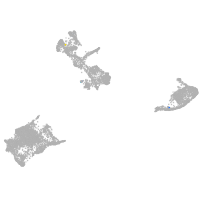 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| clic2 | 0.304 | si:dkey-194e6.1 | -0.128 |
| timp4.1 | 0.297 | acy3.2 | -0.126 |
| serpine2 | 0.294 | slc26a1 | -0.119 |
| pros1 | 0.280 | aldob | -0.118 |
| anxa5b | 0.278 | pdzk1ip1 | -0.117 |
| BX323087.1 | 0.272 | pdzk1 | -0.115 |
| slc38a8b | 0.261 | ahcyl1 | -0.115 |
| notum1b | 0.260 | dab2 | -0.111 |
| CT573484.1 | 0.255 | grhprb | -0.111 |
| CU459094.2 | 0.255 | fbp1b | -0.108 |
| tubb6 | 0.252 | cubn | -0.107 |
| gstm.3 | 0.250 | nit1 | -0.106 |
| AL954322.2 | 0.249 | agxta | -0.104 |
| anxa1a | 0.245 | pnp6 | -0.103 |
| lgals3b | 0.245 | slc26a6 | -0.102 |
| BX511100.5 | 0.244 | ftcd | -0.102 |
| olfml3b | 0.243 | upb1 | -0.101 |
| zgc:153151 | 0.236 | lrpap1 | -0.101 |
| phyhip | 0.236 | zgc:101040 | -0.101 |
| myh7l | 0.235 | gamt | -0.101 |
| emid1 | 0.231 | gstt1a | -0.101 |
| tbx2b | 0.230 | hao1 | -0.100 |
| gdpd3a | 0.229 | hoga1 | -0.100 |
| spock2 | 0.227 | alpl | -0.099 |
| kctd12b | 0.227 | fam84a | -0.099 |
| gcnt3 | 0.226 | zgc:92040 | -0.097 |
| cd151 | 0.226 | aspa | -0.097 |
| grid1a | 0.223 | lrp2a | -0.096 |
| zmat4b | 0.223 | dpydb | -0.096 |
| fkbp1b | 0.222 | dpys | -0.096 |
| wt1a | 0.221 | gnpda1 | -0.094 |
| cxcl20 | 0.221 | suclg2 | -0.093 |
| cavin2b | 0.220 | aqp8b | -0.093 |
| lmx1bb | 0.218 | si:dkey-28n18.9 | -0.093 |
| prrg2 | 0.215 | slc2a2 | -0.092 |