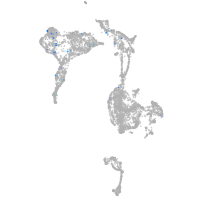
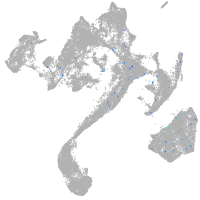
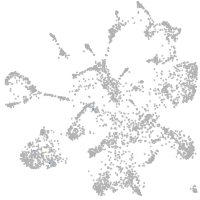
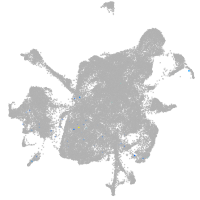
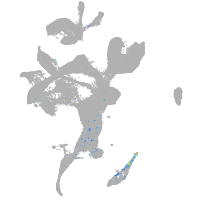
zgc:193690
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mipa | 0.253 | ptmab | -0.123 |
| crygmx | 0.245 | hmgn6 | -0.095 |
| crybb1l1 | 0.243 | CR383676.1 | -0.090 |
| crybb1l2 | 0.241 | ckbb | -0.083 |
| mipb | 0.239 | si:ch73-1a9.3 | -0.082 |
| lim2.4 | 0.238 | ptmaa | -0.072 |
| cryba1b | 0.235 | h3f3c | -0.068 |
| cryba2a | 0.234 | gpm6aa | -0.062 |
| cryba1l1 | 0.234 | gpm6ab | -0.061 |
| cryba4 | 0.232 | rnasekb | -0.059 |
| crygn2 | 0.231 | ubc | -0.059 |
| cryba2b | 0.228 | hmgb1a | -0.059 |
| crybb1 | 0.227 | tuba1c | -0.058 |
| crybb1l3 | 0.227 | atp6v0cb | -0.055 |
| lim2.3 | 0.217 | snap25b | -0.053 |
| si:ch211-255g12.6 | 0.210 | btg1 | -0.050 |
| capn3a | 0.207 | si:ch1073-429i10.3.1 | -0.049 |
| cryba1l2 | 0.206 | rtn1a | -0.049 |
| crygm2d13 | 0.200 | syt5b | -0.048 |
| grifin | 0.200 | hmgb3a | -0.048 |
| crygm2d8 | 0.198 | atp6v1g1 | -0.048 |
| lim2.5 | 0.198 | epb41a | -0.047 |
| tubb6 | 0.194 | jpt1b | -0.047 |
| gpx9 | 0.191 | atpv0e2 | -0.047 |
| cx23 | 0.190 | atp6v1e1b | -0.046 |
| crygm2d10 | 0.189 | ndrg4 | -0.046 |
| lim2.1 | 0.187 | h2afva | -0.046 |
| BFSP1 | 0.186 | histh1l | -0.045 |
| endou2 | 0.183 | atp1b2a | -0.045 |
| otc | 0.181 | crx | -0.044 |
| gja3 | 0.178 | nova2 | -0.044 |
| BX545855.1 | 0.177 | stmn1b | -0.044 |
| crygn1 | 0.177 | sypb | -0.044 |
| crygm2d12 | 0.172 | anp32e | -0.044 |
| prx | 0.171 | foxg1b | -0.043 |