zgc:162730
ZFIN 
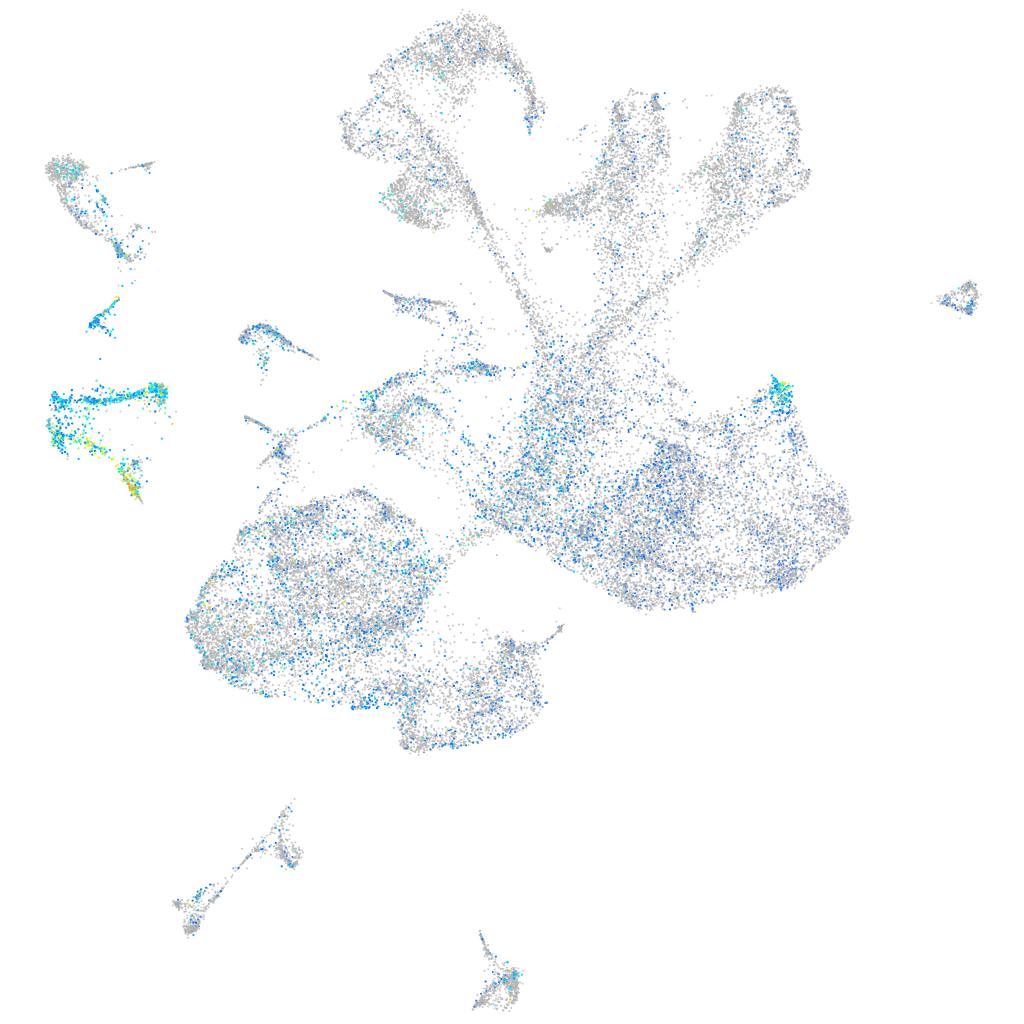
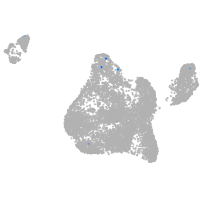
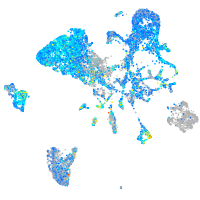
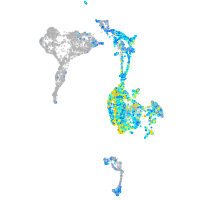
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slit1b | 0.379 | elavl3 | -0.143 |
| clu | 0.374 | ptmaa | -0.127 |
| myl9a | 0.368 | ptmab | -0.120 |
| shhb | 0.366 | hmgb3a | -0.117 |
| ctgfa | 0.361 | myt1b | -0.116 |
| npr3 | 0.361 | rtn1a | -0.113 |
| arxa | 0.354 | tubb2b | -0.107 |
| col9a3 | 0.333 | tmsb | -0.105 |
| spon1b | 0.332 | gng3 | -0.104 |
| col8a1a | 0.327 | stmn1b | -0.104 |
| shha | 0.319 | stx1b | -0.104 |
| vasna | 0.317 | marcksb | -0.104 |
| trabd2b | 0.307 | si:dkey-276j7.1 | -0.103 |
| aplnr2 | 0.307 | stxbp1a | -0.103 |
| scospondin | 0.305 | ywhah | -0.102 |
| fxyd1 | 0.304 | hnrnpaba | -0.100 |
| slit2 | 0.304 | cplx2 | -0.099 |
| optc | 0.303 | elavl4 | -0.099 |
| krt8 | 0.288 | gng2 | -0.096 |
| ptgs2a | 0.287 | onecut1 | -0.095 |
| foxa1 | 0.286 | sncb | -0.095 |
| lcn15 | 0.284 | epb41a | -0.092 |
| sulf1 | 0.284 | rtn1b | -0.092 |
| cyr61l2 | 0.281 | snap25a | -0.092 |
| si:dkey-208c12.2 | 0.279 | tmeff1b | -0.091 |
| loxl5b | 0.278 | vamp2 | -0.091 |
| col9a2 | 0.270 | tuba1c | -0.090 |
| ntn1b | 0.264 | hmgb1b | -0.090 |
| BX908782.3 | 0.259 | tmem59l | -0.088 |
| anxa4 | 0.258 | stmn2a | -0.087 |
| col9a1b | 0.256 | si:ch211-288g17.3 | -0.087 |
| tbata | 0.255 | csdc2a | -0.087 |
| sparc | 0.251 | LOC100537384 | -0.086 |
| krt18a.1 | 0.251 | nova1 | -0.086 |
| col5a1 | 0.249 | nsg2 | -0.086 |