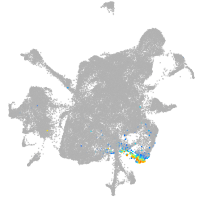
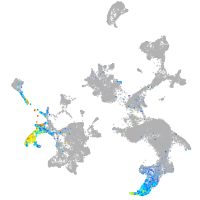
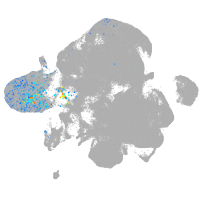
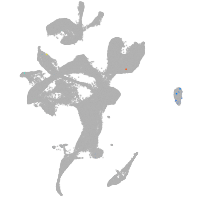
wu:fb97g03
ZFIN 
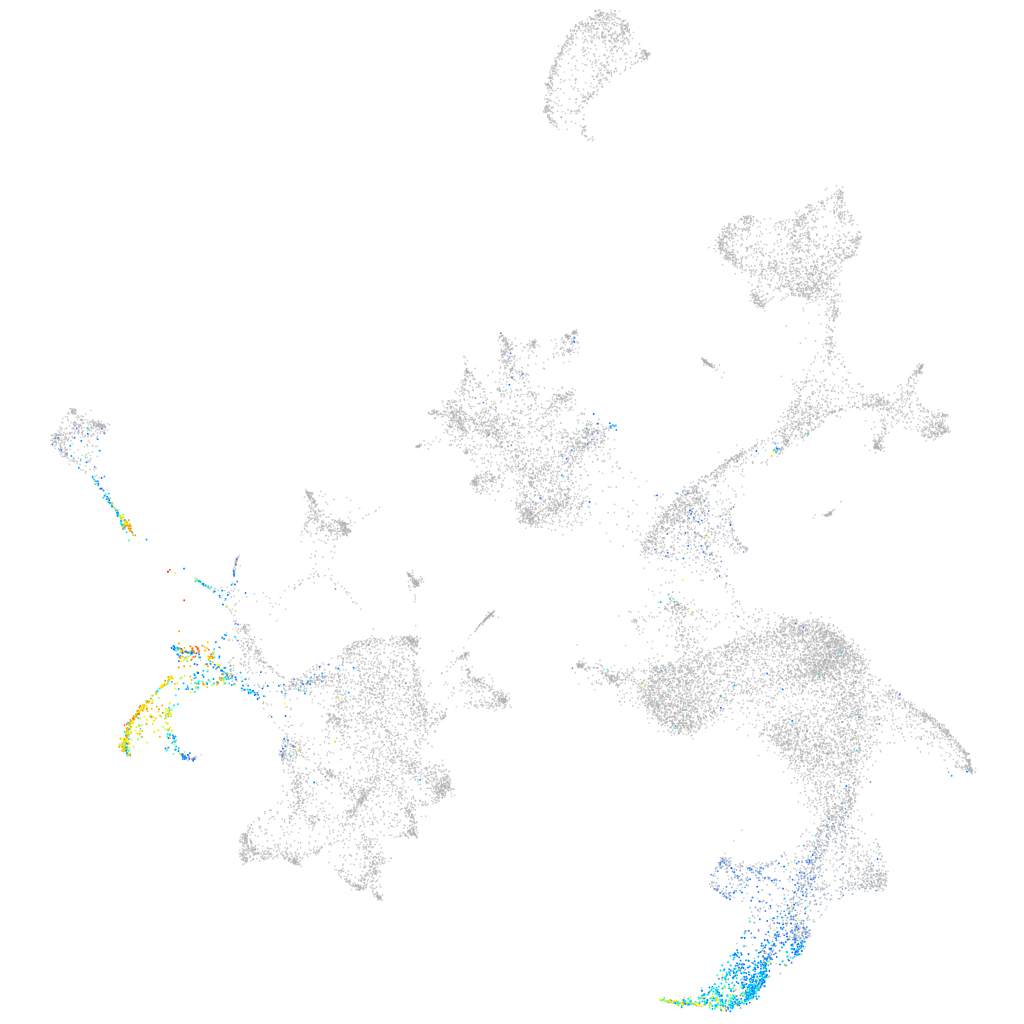
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| morc3b | 0.594 | pvalb1 | -0.214 |
| si:dkey-261j4.4 | 0.579 | pvalb2 | -0.204 |
| cldng | 0.549 | gpx1a | -0.190 |
| cpox | 0.549 | ccni | -0.187 |
| klf17 | 0.526 | gapdhs | -0.185 |
| gfi1aa | 0.523 | actc1b | -0.171 |
| znfl2a | 0.520 | cotl1 | -0.162 |
| drl | 0.516 | ptmaa | -0.159 |
| si:dkey-261j4.3 | 0.497 | si:ch211-250g4.3 | -0.151 |
| hmbsa | 0.484 | bloc1s6 | -0.143 |
| si:ch73-299h12.2 | 0.483 | srgn | -0.142 |
| bcas2 | 0.455 | mylpfa | -0.136 |
| XLOC-001964 | 0.453 | cd63 | -0.135 |
| blf | 0.449 | eno3 | -0.135 |
| snu13b | 0.437 | gabarapa | -0.129 |
| tal1 | 0.433 | arhgdig | -0.128 |
| ptges3b | 0.430 | arpc1b | -0.127 |
| npm1a | 0.421 | aldh9a1a.1 | -0.124 |
| fgf10b | 0.420 | tmsb4x | -0.123 |
| BX927258.1 | 0.409 | vat1 | -0.123 |
| mrto4 | 0.409 | crip1 | -0.121 |
| emg1 | 0.408 | ckbb | -0.119 |
| cnbpa | 0.405 | mylz3 | -0.118 |
| nop58 | 0.403 | si:ch211-260e23.9 | -0.115 |
| rcor2 | 0.402 | coro1a | -0.113 |
| sytl1 | 0.400 | tpi1b | -0.113 |
| nhp2 | 0.398 | sod1 | -0.112 |
| abce1 | 0.396 | laptm5 | -0.112 |
| setx | 0.395 | ak1 | -0.112 |
| bxdc2 | 0.394 | tmod4 | -0.112 |
| fbl | 0.386 | mdh1aa | -0.111 |
| tsr2 | 0.384 | creg1 | -0.111 |
| tma16 | 0.384 | ndrg2 | -0.111 |
| lyar | 0.382 | nfkbiab | -0.110 |
| rpl7l1 | 0.381 | zgc:162730 | -0.110 |