si:dkey-261j4.3 [Source:ZFIN;Acc:ZDB-GENE-060531-124]
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs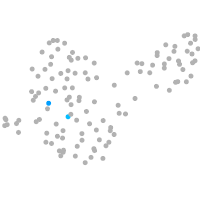 |
endoderm |
axial mesoderm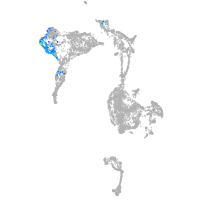 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 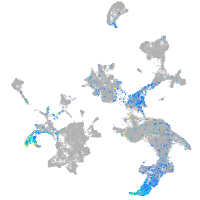 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin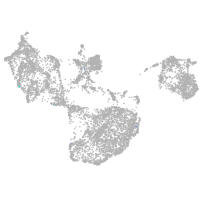 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| drl | 0.406 | tuba1c | -0.090 |
| si:dkey-261j4.4 | 0.375 | gpm6aa | -0.081 |
| blf | 0.345 | rtn1a | -0.080 |
| cldng | 0.332 | marcksl1a | -0.080 |
| si:dkey-261j4.5 | 0.321 | nova2 | -0.078 |
| si:ch73-299h12.2 | 0.307 | ccni | -0.077 |
| znfl2a | 0.285 | gapdhs | -0.077 |
| gata1a | 0.272 | mdkb | -0.077 |
| wu:fb97g03 | 0.269 | rnasekb | -0.077 |
| urod | 0.259 | atp6v0cb | -0.074 |
| hdr | 0.246 | stmn1b | -0.073 |
| susd1 | 0.246 | krt4 | -0.072 |
| tbx16 | 0.230 | elavl3 | -0.071 |
| cth1 | 0.229 | fabp3 | -0.068 |
| hmbsa | 0.215 | gng3 | -0.068 |
| si:dkey-68o6.5 | 0.212 | gpm6ab | -0.068 |
| nanog | 0.208 | cyt1 | -0.067 |
| rfesd | 0.206 | sncb | -0.066 |
| hmbsb | 0.203 | fam168a | -0.065 |
| h1m | 0.202 | si:ch1073-429i10.3.1 | -0.065 |
| klf17 | 0.201 | vamp2 | -0.064 |
| tfr1a | 0.201 | hmgb3a | -0.063 |
| cpox | 0.200 | pvalb1 | -0.063 |
| si:ch73-248e21.7 | 0.197 | pvalb2 | -0.063 |
| XLOC-020187 | 0.197 | ckbb | -0.062 |
| ddx4 | 0.196 | h2afy2 | -0.062 |
| morc3b | 0.196 | cyt1l | -0.061 |
| ccna1 | 0.195 | cspg5a | -0.060 |
| hbbe3 | 0.192 | tuba1a | -0.060 |
| fech | 0.187 | ywhag2 | -0.060 |
| gfi1b | 0.187 | elavl4 | -0.059 |
| zgc:173742 | 0.186 | atpv0e2 | -0.058 |
| si:dkeyp-114g9.1 | 0.184 | hmgn3 | -0.058 |
| zgc:163057 | 0.182 | stx1b | -0.058 |
| ikzf1 | 0.181 | tmeff1b | -0.058 |




