WT1 transcription factor a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 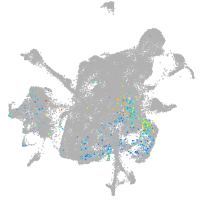 |
hematopoietic / vasculature  |
pronephros 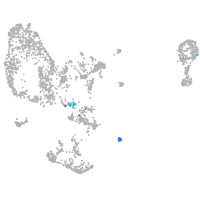 |
neural |
spinal cord / glia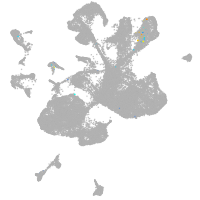 |
eye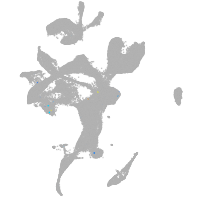 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.288 | fibina | -0.086 |
| wt1b | 0.232 | col9a1a | -0.076 |
| jam2b | 0.190 | col9a2 | -0.074 |
| meis3 | 0.189 | col2a1b | -0.074 |
| tmem88b | 0.184 | rarga | -0.073 |
| rbpms2b | 0.184 | col2a1a | -0.073 |
| tmem88a | 0.173 | col9a3 | -0.071 |
| fat2 | 0.172 | fmoda | -0.071 |
| cxcl8b.1 | 0.168 | cthrc1a | -0.070 |
| aldh1a2 | 0.168 | sox9a | -0.068 |
| znf1004 | 0.167 | tgfbi | -0.065 |
| efnb3a | 0.163 | fgfbp2b | -0.065 |
| sostdc1a | 0.162 | cnmd | -0.062 |
| cavin2b | 0.160 | six1a | -0.061 |
| gcga | 0.153 | mab21l2 | -0.060 |
| myrf | 0.149 | ecrg4a | -0.059 |
| crabp1b | 0.146 | phlda3 | -0.059 |
| nphs2 | 0.144 | CU929237.1 | -0.057 |
| cxcl8b.3 | 0.142 | dlx5a | -0.056 |
| tmem98 | 0.141 | col11a2 | -0.055 |
| naalad2 | 0.138 | LO018188.1 | -0.055 |
| kirrel1b | 0.138 | fxyd1 | -0.052 |
| agrp | 0.134 | zgc:153675 | -0.051 |
| itgb6 | 0.130 | eya1 | -0.051 |
| ptprq | 0.129 | fkbp11 | -0.050 |
| krt94 | 0.128 | VIT | -0.050 |
| pcdh10b | 0.127 | matn4 | -0.050 |
| lurap1 | 0.127 | ahcy | -0.049 |
| bcam | 0.124 | bhmt | -0.048 |
| ftr82 | 0.121 | kdelr3 | -0.047 |
| alkal2b | 0.120 | lpar1 | -0.046 |
| lama5 | 0.119 | tnc | -0.046 |
| wnt7bb | 0.119 | six2a | -0.046 |
| hoxc8a | 0.119 | col11a1b | -0.046 |
| tmem108 | 0.116 | dlx2a | -0.045 |




