WT1 transcription factor a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 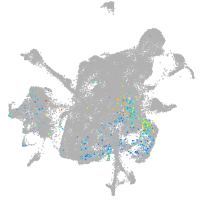 |
hematopoietic / vasculature  |
pronephros 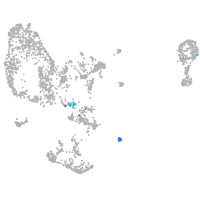 |
neural |
spinal cord / glia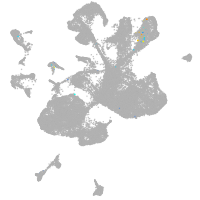 |
eye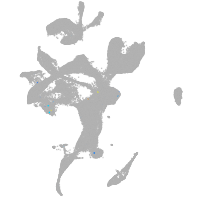 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| meis3 | 0.237 | cycsb | -0.093 |
| pcolcea | 0.232 | hsp90aa1.1 | -0.072 |
| tmem88a | 0.227 | atp5f1e | -0.071 |
| krt94 | 0.222 | hspe1 | -0.069 |
| alx1 | 0.214 | ttn.1 | -0.069 |
| tmem88b | 0.212 | cox7b | -0.067 |
| emx2 | 0.210 | hspb1 | -0.067 |
| crabp1b | 0.201 | ttn.2 | -0.063 |
| ccl25b | 0.192 | zgc:92429 | -0.063 |
| meis2b | 0.187 | tmem38a | -0.062 |
| tnfsf10 | 0.186 | uqcrh | -0.062 |
| si:ch211-286o17.1 | 0.178 | klhl41b | -0.062 |
| smoc2 | 0.176 | pabpc4 | -0.061 |
| sostdc1a | 0.175 | chchd10 | -0.061 |
| bmp16 | 0.172 | cox5aa | -0.061 |
| dhrs3a | 0.165 | fxr2 | -0.061 |
| tpm4a | 0.156 | cox7c | -0.060 |
| rpz5 | 0.152 | desma | -0.060 |
| pbx1b | 0.152 | aldoab | -0.060 |
| rbp5 | 0.150 | c1qbp | -0.060 |
| zgc:162612 | 0.149 | uqcrfs1 | -0.060 |
| igf2b | 0.149 | actc1a | -0.059 |
| zfp36l1a | 0.148 | acta1a | -0.059 |
| cnn3a | 0.148 | hspd1 | -0.058 |
| cald1a | 0.147 | unc45b | -0.058 |
| pbx1a | 0.145 | mybphb | -0.058 |
| zgc:154093 | 0.145 | cox6c | -0.057 |
| ppfibp1a | 0.143 | srl | -0.057 |
| ftr82 | 0.141 | atp2a1 | -0.057 |
| jam2b | 0.141 | myod1 | -0.057 |
| cdh11 | 0.139 | ldb3a | -0.057 |
| pmp22a | 0.139 | gatm | -0.057 |
| podxl | 0.138 | gapdh | -0.057 |
| tmem108 | 0.134 | cox7a2a | -0.056 |
| pdgfra | 0.132 | ak1 | -0.056 |




