WT1 transcription factor a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 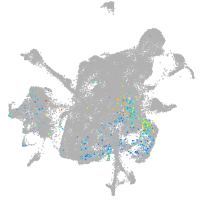 |
hematopoietic / vasculature  |
pronephros 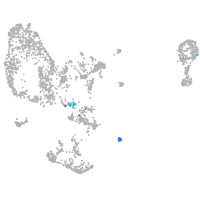 |
neural |
spinal cord / glia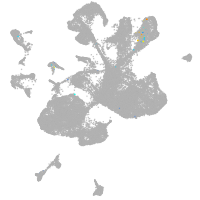 |
eye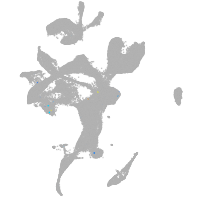 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.350 | BX088707.3 | -0.195 |
| krt94 | 0.333 | acta2 | -0.172 |
| jam2b | 0.293 | tpm1 | -0.169 |
| krt15 | 0.266 | mylkb | -0.167 |
| tmem88b | 0.256 | tagln | -0.166 |
| bcam | 0.247 | lmod1b | -0.150 |
| gstm.3 | 0.238 | myl9a | -0.148 |
| si:ch211-156j16.1 | 0.237 | myl6 | -0.147 |
| phactr4a | 0.232 | myh11a | -0.137 |
| krt8 | 0.220 | tpm2 | -0.133 |
| cav1 | 0.219 | csrp1b | -0.130 |
| ftr82 | 0.216 | tmem119b | -0.128 |
| cavin2b | 0.212 | sat1a.2 | -0.128 |
| cd151 | 0.211 | cald1b | -0.127 |
| aqp1a.1 | 0.209 | inka1a | -0.123 |
| fgf24 | 0.198 | gapdhs | -0.123 |
| scarb2a | 0.196 | desmb | -0.121 |
| dag1 | 0.195 | cnn1b | -0.121 |
| bnc2 | 0.195 | gpia | -0.119 |
| col12a1a | 0.194 | myocd | -0.117 |
| cxcl8b.1 | 0.192 | ckba | -0.114 |
| tbx18 | 0.185 | mdkb | -0.114 |
| aldh1a2 | 0.184 | foxf2a | -0.112 |
| hspb1 | 0.181 | ckbb | -0.112 |
| lurap1 | 0.180 | cfl1 | -0.111 |
| boc | 0.179 | gng2 | -0.110 |
| tspan18a | 0.176 | eno3 | -0.110 |
| nrp1a | 0.175 | itga1 | -0.109 |
| cavin1b | 0.173 | calm2a | -0.107 |
| bmper | 0.173 | tuba8l2 | -0.107 |
| palm1b | 0.171 | cox6b1 | -0.105 |
| eppk1 | 0.169 | fhl1a | -0.104 |
| tmem88a | 0.166 | vdac1 | -0.102 |
| mmel1 | 0.161 | angptl5 | -0.101 |
| stx2b | 0.161 | tpi1b | -0.101 |




