WNT1 inducible signaling pathway protein 3 [Source:ZFIN;Acc:ZDB-GENE-041001-86]
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 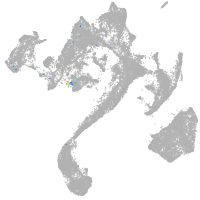 |
mural cells / non-skeletal muscle 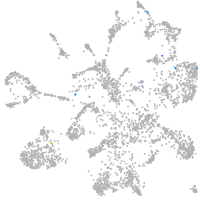 |
mesenchyme 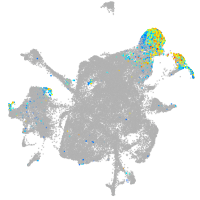 |
hematopoietic / vasculature 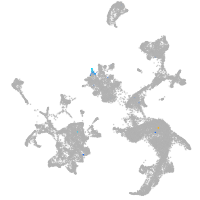 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm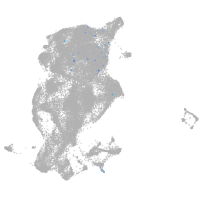 |
fin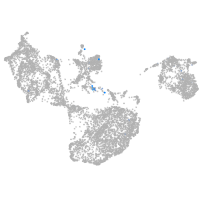 |
ionocytes / mucous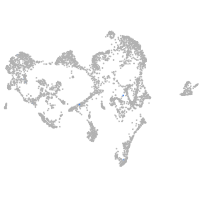 |
pigment cells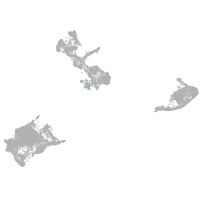 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110440032 | 0.422 | arl6ip1 | -0.037 |
| cxcl8b.3 | 0.382 | cldnb | -0.031 |
| nmbr | 0.338 | lsm8 | -0.031 |
| cmlc1 | 0.285 | egr1 | -0.029 |
| acanb | 0.273 | bik | -0.028 |
| itih1 | 0.270 | tprg1 | -0.028 |
| emilin3a | 0.263 | psmc1a | -0.027 |
| LOC793010 | 0.261 | gbp | -0.026 |
| tbx5a | 0.261 | sypl2a | -0.025 |
| rx1 | 0.260 | fthl27 | -0.025 |
| loxl1 | 0.236 | gbgt1l4 | -0.025 |
| si:dkey-6n6.1 | 0.232 | rhebl1 | -0.025 |
| acana | 0.229 | prrg2 | -0.025 |
| col9a1a | 0.227 | larp4ab | -0.025 |
| zgc:101100 | 0.225 | mapkapk2a | -0.025 |
| f2rl2 | 0.218 | gcsha | -0.025 |
| matn1 | 0.208 | ube2h | -0.025 |
| rgs14a | 0.203 | vapal | -0.024 |
| enpp7.1 | 0.199 | hnrnpl2 | -0.024 |
| coch | 0.198 | tfb2m | -0.024 |
| meltf | 0.198 | ptbp1a | -0.024 |
| CR456642.1 | 0.197 | uqcc1 | -0.024 |
| mia | 0.190 | ammecr1 | -0.024 |
| BX005421.1 | 0.189 | bin3 | -0.023 |
| col8a2 | 0.189 | zgc:101783 | -0.023 |
| otos | 0.189 | flot1b | -0.023 |
| XLOC-000910 | 0.189 | stx12 | -0.023 |
| hapln1b | 0.188 | fis1 | -0.023 |
| clcn1b | 0.187 | ppig | -0.023 |
| LOC101882950 | 0.185 | c1qbp | -0.023 |
| fgfbp2b | 0.170 | rpz2 | -0.023 |
| hoxd13a | 0.168 | hk1 | -0.023 |
| ENKD1 | 0.167 | uri1 | -0.023 |
| col9a2 | 0.167 | cdk7 | -0.023 |
| LOC103909344 | 0.160 | grhl1 | -0.022 |