regulator of G protein signaling 14a
ZFIN 
Other cell groups


all cells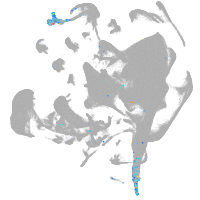 |
blastomeres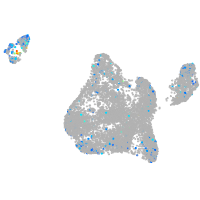 |
PGCs |
endoderm |
axial mesoderm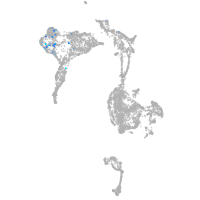 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 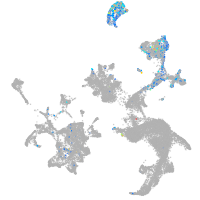 |
pronephros 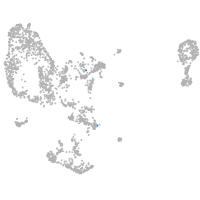 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous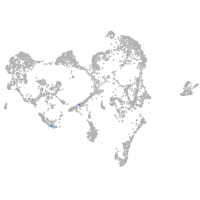 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctss2.1 | 0.324 | tuba1c | -0.046 |
| glipr1a | 0.311 | hmgb3a | -0.044 |
| laptm5 | 0.309 | nova2 | -0.042 |
| si:dkey-102g19.3 | 0.304 | gpm6aa | -0.041 |
| spi1b | 0.304 | mdkb | -0.039 |
| coro1a | 0.298 | rtn1a | -0.039 |
| plek | 0.298 | rnasekb | -0.038 |
| myo1f | 0.291 | tuba1a | -0.037 |
| ncf1 | 0.290 | atp6v0cb | -0.036 |
| itgb2 | 0.289 | fabp3 | -0.036 |
| alox5ap | 0.287 | tmeff1b | -0.036 |
| si:ch211-193e13.5 | 0.287 | elavl3 | -0.035 |
| si:dkey-27h10.2 | 0.287 | fam168a | -0.035 |
| grap2b | 0.285 | gpm6ab | -0.035 |
| fcer1gl | 0.284 | hmgb1b | -0.035 |
| tnfaip8l2a | 0.280 | stmn1b | -0.035 |
| lcp1 | 0.278 | h1f0 | -0.032 |
| wasa | 0.277 | krt4 | -0.032 |
| wasb | 0.277 | si:ch211-137a8.4 | -0.032 |
| nrros | 0.275 | cspg5a | -0.031 |
| ptprc | 0.272 | gng3 | -0.031 |
| cfbl | 0.271 | mdka | -0.031 |
| si:ch73-248e21.5 | 0.271 | cyt1 | -0.030 |
| lpar5a | 0.271 | h2afy2 | -0.030 |
| hmha1b | 0.266 | si:ch73-46j18.5 | -0.029 |
| FERMT3 (1 of many) | 0.266 | sncb | -0.029 |
| parvg | 0.265 | tmsb | -0.029 |
| si:ch211-243a20.4 | 0.264 | tubb5 | -0.029 |
| itgae.2 | 0.263 | vamp2 | -0.029 |
| scpp8 | 0.262 | calm1a | -0.029 |
| si:ch211-276a23.5 | 0.262 | meis1b | -0.028 |
| gpr183a | 0.261 | cadm3 | -0.027 |
| sla1 | 0.261 | cyt1l | -0.027 |
| si:dkey-53k12.2 | 0.261 | epb41a | -0.027 |
| CU499336.2 | 0.260 | rtn1b | -0.027 |




