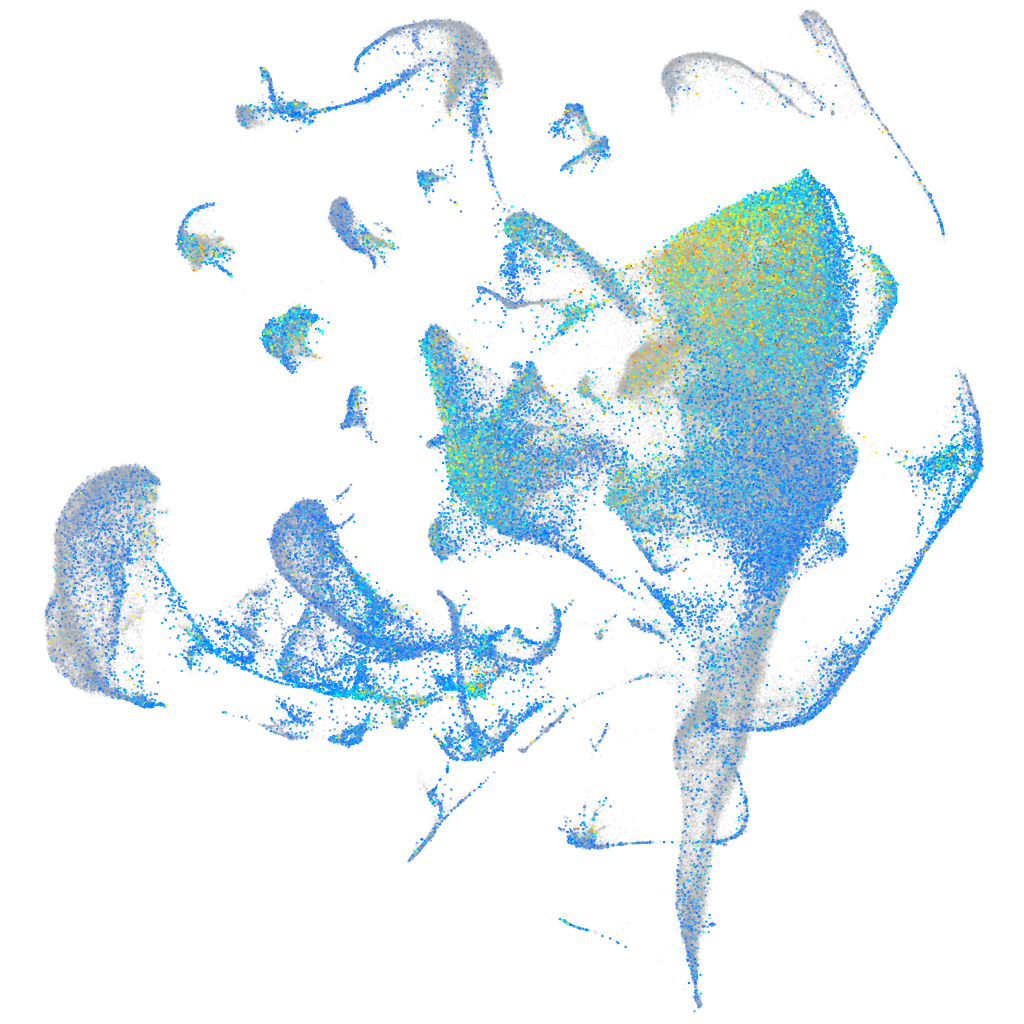rapunzel 2
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:65894 | 0.144 | mki67 | -0.082 |
| sncb | 0.143 | lig1 | -0.077 |
| ywhag2 | 0.143 | zgc:110425 | -0.073 |
| gng3 | 0.142 | ccna2 | -0.072 |
| calm1a | 0.140 | mibp | -0.072 |
| snap25a | 0.137 | tpx2 | -0.072 |
| stmn2a | 0.137 | chaf1a | -0.071 |
| atp6v1e1b | 0.136 | fbxo5 | -0.070 |
| calm1b | 0.136 | si:dkey-66i24.9 | -0.069 |
| vamp2 | 0.134 | zgc:110216 | -0.069 |
| gapdhs | 0.131 | ccnb1 | -0.068 |
| atp6v1g1 | 0.130 | dlgap5 | -0.068 |
| stxbp1a | 0.130 | hspb1 | -0.068 |
| rnasekb | 0.129 | banf1 | -0.067 |
| atp6v0cb | 0.128 | dek | -0.067 |
| calm2a | 0.127 | cdca8 | -0.065 |
| map1aa | 0.126 | mad2l1 | -0.065 |
| stmn1b | 0.125 | pou5f3 | -0.065 |
| mllt11 | 0.124 | si:ch211-113a14.18 | -0.065 |
| si:dkeyp-75h12.5 | 0.124 | si:dkey-108k21.10 | -0.065 |
| scg2b | 0.123 | smc4 | -0.065 |
| eno2 | 0.119 | stm | -0.065 |
| gabarapb | 0.118 | nusap1 | -0.063 |
| cdc42 | 0.117 | ube2c | -0.063 |
| brk1 | 0.116 | hnrnpa1b | -0.062 |
| gnb1a | 0.116 | kmt5ab | -0.060 |
| hsbp1a | 0.116 | pcna | -0.060 |
| rab6bb | 0.116 | rrm1 | -0.060 |
| syn2a | 0.116 | zgc:153405 | -0.060 |
| uchl1 | 0.116 | zmp:0000000624 | -0.059 |
| calm3b | 0.115 | aep1 | -0.058 |
| camk2n1a | 0.115 | ccnd1 | -0.058 |
| hist2h2l | 0.115 | COX7A2 | -0.058 |
| olfm1b | 0.115 | si:ch211-161h7.4 | -0.058 |
| atp6ap2 | 0.114 | slbp | -0.058 |