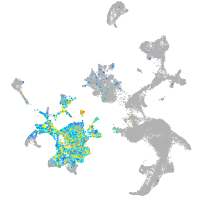
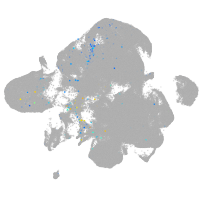
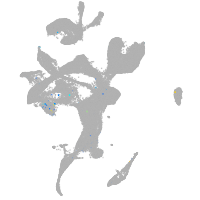
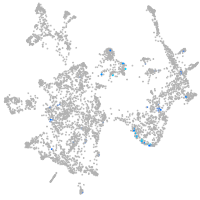
transmembrane protein 88 a
ZFIN 
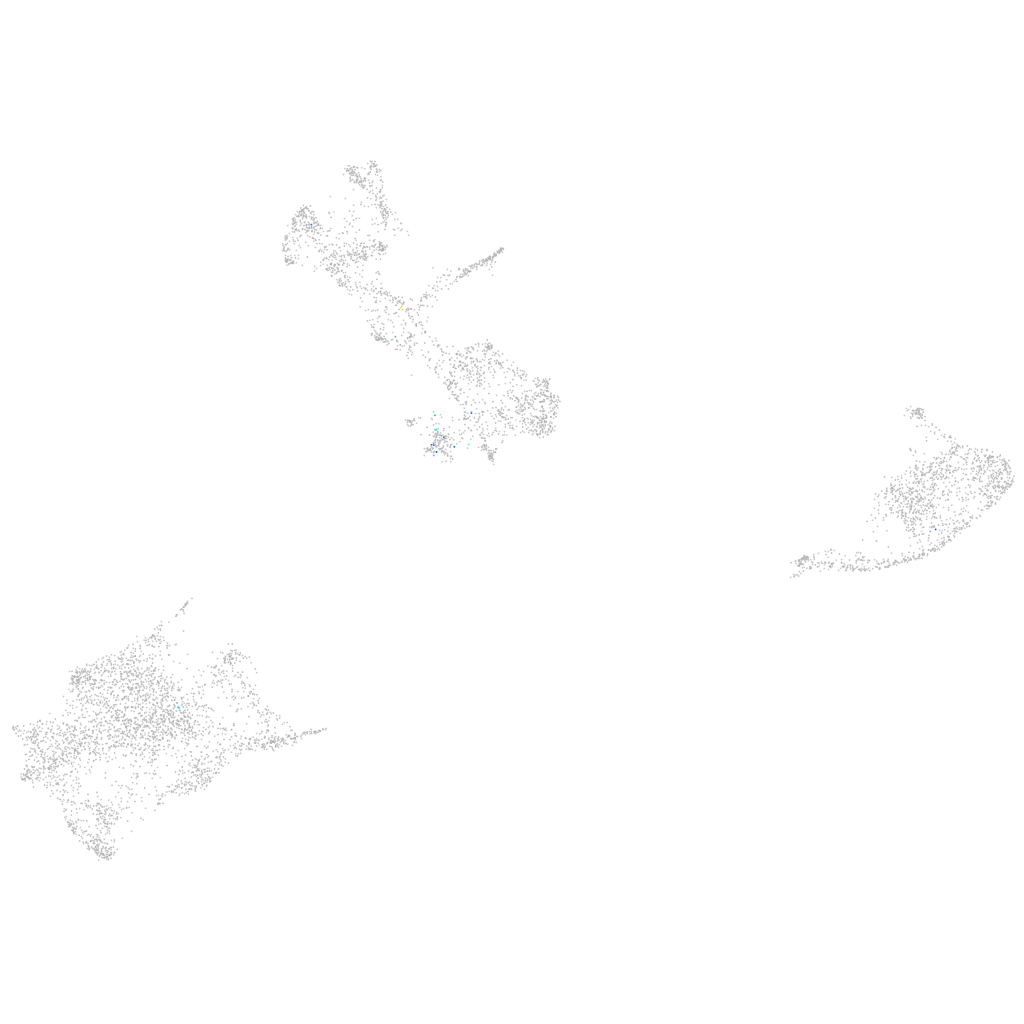
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX936342.1 | 0.374 | pfas | -0.026 |
| CABZ01111454.1 | 0.331 | cyt1 | -0.025 |
| CT573429.1 | 0.316 | marcksl1b | -0.024 |
| calcoco1b | 0.311 | slc2a15a | -0.024 |
| CR759846.1 | 0.289 | prdx6 | -0.023 |
| lrrc53 | 0.272 | LOC103910009 | -0.022 |
| tie1 | 0.256 | rab27a | -0.022 |
| BX927219.1 | 0.250 | hbae3 | -0.022 |
| si:ch211-33e4.2 | 0.250 | zgc:110339 | -0.021 |
| vipr1a | 0.238 | uqcrh | -0.021 |
| kdr | 0.238 | atox1 | -0.021 |
| LOC101882264 | 0.236 | calm2b | -0.021 |
| LOC103910198 | 0.235 | prelid3b | -0.021 |
| lrrc75bb | 0.234 | vps29 | -0.020 |
| mir214a | 0.221 | reep5 | -0.020 |
| sned1 | 0.221 | si:dkey-251i10.2 | -0.020 |
| FO904980.1 | 0.220 | pgd | -0.020 |
| btr16.1 | 0.218 | tmem130 | -0.020 |
| cpa6 | 0.211 | serinc1 | -0.019 |
| myct1a | 0.209 | phlda2 | -0.019 |
| htr7c | 0.208 | bace2 | -0.019 |
| ghrh | 0.207 | phyhd1 | -0.019 |
| mmp14a | 0.205 | apoa2 | -0.018 |
| erg | 0.204 | syngr1a | -0.018 |
| si:ch211-66e2.3 | 0.201 | nme4 | -0.018 |
| LOC108180727 | 0.198 | atp5if1b | -0.018 |
| LOC100006216 | 0.196 | fabp3 | -0.018 |
| tmem88b | 0.192 | spock3 | -0.018 |
| adamts6 | 0.190 | rab38 | -0.018 |
| prdm5 | 0.190 | ndufb2 | -0.018 |
| CR925723.1 | 0.184 | MED9 | -0.018 |
| BX072576.1 | 0.183 | cmtm7 | -0.018 |
| si:ch211-136m16.8 | 0.179 | gsna | -0.018 |
| si:rp71-39b20.4 | 0.179 | vma21 | -0.017 |
| dnmt3bb.1 | 0.176 | trappc3 | -0.017 |