transmembrane protein 88 a
ZFIN 
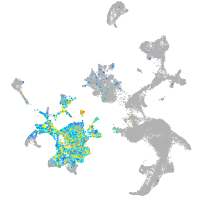
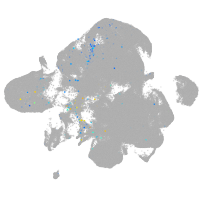
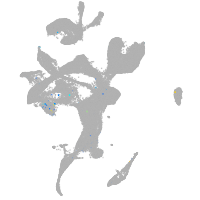
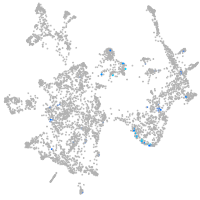
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpnmb | 0.527 | slc25a5 | -0.046 |
| LOC108180727 | 0.445 | btf3 | -0.044 |
| gypc | 0.425 | cldnb | -0.041 |
| kdrl | 0.404 | ier2b | -0.041 |
| aqp8a.1 | 0.394 | pabpc1a | -0.040 |
| adgrl4 | 0.382 | selenow1 | -0.034 |
| rasip1 | 0.377 | fosab | -0.033 |
| egfl7 | 0.368 | eef2b | -0.033 |
| camkvl | 0.367 | cldn7b | -0.031 |
| nr0b2a | 0.354 | cnpy1 | -0.031 |
| XLOC-004037 | 0.348 | wu:fb18f06 | -0.030 |
| clec14a | 0.329 | si:dkey-87o1.2 | -0.030 |
| sele | 0.321 | pycard | -0.029 |
| thsd1 | 0.316 | dap1b | -0.029 |
| XLOC-036510 | 0.303 | gnb1a | -0.029 |
| LOC100331510 | 0.301 | btg1 | -0.028 |
| cby1 | 0.294 | tmem14ca | -0.028 |
| kiaa1549la | 0.293 | BX000438.2 | -0.028 |
| aqp1a.1 | 0.291 | eif2s3 | -0.028 |
| cacna1ha | 0.291 | krt4 | -0.027 |
| mcamb | 0.283 | serinc2 | -0.027 |
| lmo2 | 0.278 | perp | -0.027 |
| FP016239.1 | 0.277 | jun | -0.026 |
| AL954322.2 | 0.275 | glrx | -0.026 |
| flt4 | 0.269 | capns1a | -0.026 |
| bicd2 | 0.268 | cebpb | -0.026 |
| myoz1a | 0.266 | fosb | -0.026 |
| esm1 | 0.262 | si:ch1073-443f11.2 | -0.026 |
| zgc:113363 | 0.260 | hdlbpa | -0.026 |
| si:ch73-116o1.2 | 0.260 | atp5f1e | -0.026 |
| hoxb6a | 0.255 | cldne | -0.026 |
| tmem54b | 0.251 | prr13 | -0.025 |
| hhex | 0.246 | si:ch73-335l21.4 | -0.025 |
| CR848746.1 | 0.243 | ola1 | -0.025 |
| ecscr | 0.243 | trappc5 | -0.024 |